
How the Scientific Python ecosystem helps answer fundamental questions of the Universe
Abstract¶
The ATLAS experiment at CERN explores vast amounts of physics data to answer the most fundamental questions of the Universe. The prevalence of Python in scientific computing motivated ATLAS to adopt it for its data analysis workflows while enhancing users’ experience. This paper will describe to a broad audience how a large scientific collaboration leverages the power of the Scientific Python ecosystem to tackle domain-specific challenges and advance our understanding of the Cosmos. Through a simplified example of the renowned Higgs boson discovery, attendees will gain insights into the utilization of Python libraries to discriminate a signal in immersive noise, through tasks such as data cleaning, feature engineering, statistical interpretation and visualization at scale.
1Introduction¶
The field of high energy physics (HEP) is devoted to the study of the fundamental forces of Nature and their interactions with matter. To study the structure of the Universe on the smallest scales requires the highest energy density environments possible — similar to those of the early Universe. These extreme energy density environments are created at the CERN international laboratory, in Geneva, Switzerland, using the Large Hadron Collider (LHC) to collide “bunches” of billions of protons at a center-of-mass energy of . The resulting collisions are recorded with building-sized particle detectors positioned around the LHC’s ring that are designed to measure subatomic particle properties. Given the rarity of the subatomic phenomena of interest, the rate of the beam crossings is a tremendous to maximize the number of high quality collisions that can be captured and read out by the detectors. Even with real-time onboard processing (“triggering”) of the experiment detector readout to save only the most interesting collisions, detectors like the ATLAS experiment ATLAS Collaboration, 2008 still produce multiple petabytes of data per year. These data are then further filtered through selection criteria on the topology and kinematic quantities of the particle collision “events” recorded into specialized datasets for different kinds of physics analysis. The final datasets that physicists use in their physics analyses in ATLAS is still on the order of hundreds of terabytes, which poses challenges of compute scale and analyst time to efficiently use while maximizing physics value.
Traditionally, the ATLAS and the other LHC experiment have created experiment-specific custom C++ frameworks to handle all stages of the data processing pipeline, from the initial construction of high-level physics objects from the raw data to the final statistical analyses.
Motivated by the broad success of the Scientific Python ecosystem across many domains of science, and the rise of the Scikit-HEP ecosystem of Pythonic tooling for particle physics Rodrigues & others, 2020Schreiner et al., 2022 and community tools produced by the Institute for Research and Innovation in Software for High Energy Physics (IRIS-HEP) Elmer et al., 2017Albrecht & others, 2019, there has been a broad community-driven shift in HEP towards use of the Scientific Python ecosystem for analysis of physics data — a PyHEP ecosystem HEP Software Foundation, 2024.
The use of dataframes and array programming for data analysis has enhanced the user experience while providing efficient computations without the need of coding optimized low-level routines.
The ATLAS collaboration is further extending this ecosystem of tooling to include high-level custom Python bindings to the low level C++ frameworks using nanobind Jakob, 2022.
Collectively, these tools are modernizing the methods which researchers are engaging data analysis at large scale and providing a novel end-to-end analysis ecosystem for the ATLAS collaboration.
2Employing the Scientific Python ecosystem¶
The multiple stages of physics data processing and analysis map onto different parts of the Scientific Python ecosystem.
This begins with the highly-structured but jagged nature of the event data in HEP.
The data structure of each event consists of variable length lists of physics objects (e.g. electrons, collections of tracks from charged objects).
To study the properties of the physics objects in a statistical manner, a fixed event analysis procedure is repeated over billions of events.
This has traditionally motivated the use of “event loops” that implicitly construct event-level quantities of interest and leveraged the C++ compiler to produce efficient iterative code.
This precedent made it difficult to take advantage of array programming paradigms that are common in Scientific Python given NumPy Harris et al., 2020 vector operations.
The Scikit-HEP library Awkward Array Pivarski et al., 2018 provides a path forward by providing NumPy-like idioms for nested, variable-sized (JSON-like) data and also brings analysts into an array programming paradigm Hartmann et al., 2021.
With the ability to operate on HEP data structures in an array programming — or “columnar” — approach, the next step is to be able to read and write with the HEP domain specific ROOT Brun & Rademakers, 1997 file format — which has given the particle physics community columnar data structures with efficient compression since 1997 Pivarski et al., 2020.
This is accomplished with use of the uproot library Pivarski et al., 2017, which allows for efficient transformation of ROOT data to NumPy or Awkward arrays.
The data is then filtered through kinematic and physics signature motivated selections using Awkward manipulations and queries to create array collections that contain the passing events.
Through intense detector characterization and calibration efforts, the ATLAS collaboration has developed robust methods and tooling to apply corrections to the data and evaluate systematic uncertainties.
For instance, corrections to the signal collected by a specific calorimeter subsystem along with systematic uncertainties due to the imperfect knowledge of the subsystem.
Given the custom nature of the detector and correction implementations, these corrections are implemented in custom C++ libraries in the ATLAS software framework, Athena ATLAS Collaboration, 2021ATLAS Collaboration, 2021.
To expose these C++ libraries to the Pythonic tooling layer, custom Python bindings are written using nanobind for high efficiency, as seen in Figure 1.
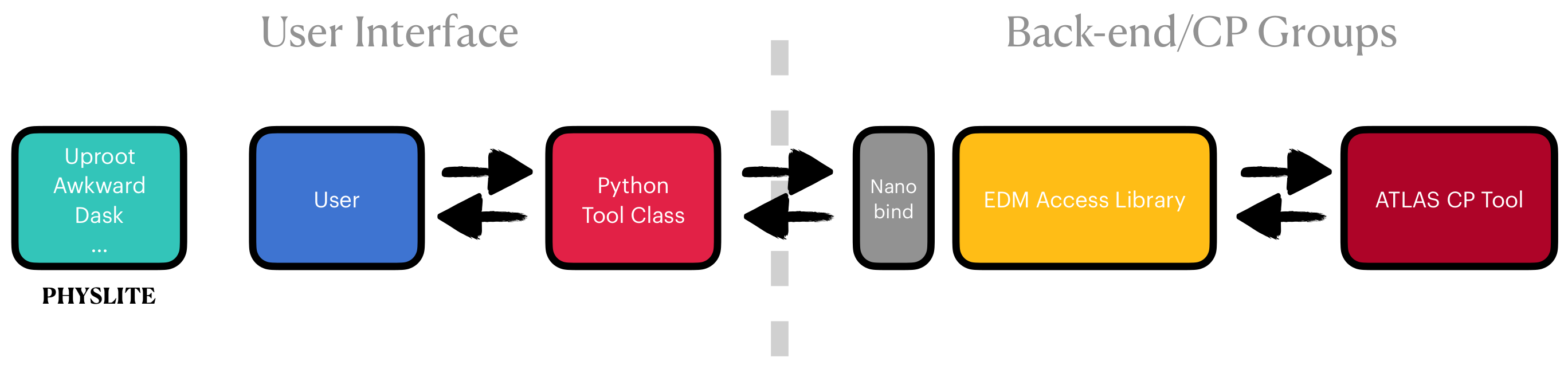
Figure 1:The data access abstract interface from the high level user facing Python API to the ATLAS Event Data Model (EDM) access library that exposes the shared ATLAS combined performance (CP) tools for reconstruction, identification, and measurement of physics objects. Kourlitis et al., 2024
To contend with the extreme data volume, efficient distributed computing is an essential requirement.
Given the success of Dask Dask Development Team, 2016 in the Scientific Python ecosystem, and its ability to be deployed across both traditional batch systems and cloud based infrastructure with Kubernetes, the Scikit-HEP ecosystem has built extensions to Dask that allow for native Dask collections of Awkward arrays dask-awkward Development Team, 2022 and computing multidimensional boost-histogram objects Schreiner et al., 2018 with Dask collections dask-histogram Development Team, 2021.
Using Dask and these extensions, the data selection and systematic correction workflow is able to be horizontally scaled out across ATLAS collaboration compute resources to provide the data throughput necessary to make analysis feasible.
This is often achieved through use of the high level coffea columnar analysis framework Gray et al., 2019 which was designed to integrate with Dask and these HEP specific Dask extensions.
The resulting data objects that are returned to analysts are histograms of physics quantity distributions — such as the reconstructed invariant-mass of a collection of particles or particle momentum.
Using the hist library Schreiner et al., n.d. for higher level data exploration and manipulation, physicists are then able to efficiently further manipulate the data distributions using tooling from the broader Scientific Python ecosystem and create domain-centric visualizations using the mplhep Novak et al., 2020 extension of Matplotlib Hunter, 2007.
From these high level data representations of the relevant physics, physicists are then able to serialize the distributions and use them for the final stages of data analysis and statistical modeling and inference.
3Uncovering the Higgs boson¶
The most famous and revolutionary discovery in particle physics this century is the discovery of the Higgs boson — the particle corresponding to the quantum field that gives mass to fundamental particles through the Brout-Englert-Higgs mechanism — by the ATLAS and CMS experimental collaborations in 2012. ATLAS Collaboration, 2012CMS Collaboration, 2012
This discovery work was done using large amounts of customized C++ software, but in the following decade the state of the PyHEP community has advanced enough that the workflow can now be done using community Python tooling.
To provide an overview of the Pythonic tooling and functionality, a high level summary of a simplified analysis workflow Held et al., 2023 of a Higgs “decay” to two intermediate bosons that decay to charged leptons (i.e. electrons () and muons (μ)), , on ATLAS open data ATLAS Collaboration, 2020 is summarized in this section.
3.1Loading data¶
Given the size of the data, the files used in a real analysis will usually be cached at a national level “analysis facility” where the analysis code will run.
Using coffea, uproot, and Dask, these files can then be efficiently read and the tree structure of the data can populate Awkward arrays.
from coffea.nanoevents import NanoEventsFactory, PHYSLITESchema
def get_uris_from_cache(): ...
def filter_name(name):
return name in (
"AnalysisElectronsAuxDyn.pt",
"AnalysisElectronsAuxDyn.eta",
"AnalysisElectronsAuxDyn.phi",
"AnalysisElectronsAuxDyn.m",
"AnalysisElectronsAuxDyn.charge",
...,
)
file_uris = get_uris_from_cache(...)
# uproot used internally to read files into Awkward arrays
events_mc = NanoEventsFactory.from_root(
file_uris,
schemaclass=PHYSLITESchema,
uproot_options=dict(filter_name=filter_name),
permit_dask=True,
).events()
Using coffea, tree structured ROOT files are read with uproot from an efficient file cache, and the relevant branches for physics are filtered out into Awkward arrays. The operation is scaled out on a Dask cluster for read performance.
3.2Cleaning and selecting data¶
Once the data is in Awkward arrays, additional selections need to be applied before it can be analyzed. Only physics objects of adequate quality are kept for further analysis and those should reconstruct the topology of interest. In this particular case, due to the decay of the Higgs boson to two leptons, the data selected contain four charged leptons grouped in two opposite flavor lepton pairs (so that the total charge is zero, as the Higgs and the -bosons are electrically neutral). Additionally, in order to compare various kinds of simulated data, the events need to be normalized/weighted given their relative appearance in reality and the amount of actual data collected by the experiment.
These selection and weighting can then be implemented in an analysis specific coffea processor, and then the processor can be executed used a Dask executor to horizontally scale out the analysis selection across the available compute.
import awkward as ak
import hist
import vector
from coffea import processor
from distributed import Client
def get_xsec_weight(sample, infofile):
"""Returns normalization weight for a given sample."""
lumi = 10_000 # pb^-1
xsec_map = infofile.infos[sample] # dictionary with event weighting information
xsec_weight = (lumi * xsec_map["xsec"]) / (xsec_map["sumw"] * xsec_map["red_eff"])
return xsec_weight
def lepton_filter(lep_charge, lep_type):
"""Filters leptons: sum of charges is required to be 0, and sum of lepton types 44/48/52.
Electrons have type 11, muons have 13, so this means 4e/4mu/2e2mu.
"""
sum_lep_charge = ak.sum(lep_charge, axis=1)
sum_lep_type = ak.sum(lep_type, axis=1)
good_lep_type = ak.any(
[sum_lep_type == 44, sum_lep_type == 48, sum_lep_type == 52], axis=0
)
return ak.all([sum_lep_charge == 0, good_lep_type], axis=0)
class HZZAnalysis(processor.ProcessorABC):
"""The coffea processor used in this analysis."""
def __init__(self):
pass
def process(self, events):
# The process function performs columnar operations on the events
# passed to it and applies all the corrections and selections to
# either the simulation or the data (e.g. get_xsec_weight and
# lepton_filter). All the event level data selection occurs here
# and returns accumulators with the selections.
vector.register_awkward()
# type of dataset being processed, provided via metadata (comes originally from fileset)
dataset_category = events.metadata["dataset_name"]
# apply a cut to events, based on lepton charge and lepton type
events = events[lepton_filter(events.lep_charge, events.lep_typeid)]
# construct lepton four-vectors
leptons = ak.zip(
{
"pt": events.lep_pt,
"eta": events.lep_eta,
"phi": events.lep_phi,
"energy": events.lep_energy,
},
with_name="Momentum4D",
)
# calculate the 4-lepton invariant mass for each remaining event
# this could also be an expensive calculation using external tools
mllll = (
leptons[:, 0] + leptons[:, 1] + leptons[:, 2] + leptons[:, 3]
).mass / 1000
# create histogram holding outputs, for data just binned in m4l
mllllhist_data = hist.Hist.new.Reg(
num_bins,
bin_edge_low,
bin_edge_high,
name="mllll",
label="$\mathrm{m_{4l}}$ [GeV]",
).Weight() # using weighted storage here for plotting later, but not needed
# three histogram axes for MC: m4l, category, and variation (nominal and
# systematic variations)
mllllhist_MC = (
hist.Hist.new.Reg(
num_bins,
bin_edge_low,
bin_edge_high,
name="mllll",
label="$\mathrm{m_{4l}}$ [GeV]",
)
.StrCat([k for k in fileset.keys() if k != "Data"], name="dataset")
.StrCat(
["nominal", "scaleFactorUP", "scaleFactorDOWN", "m4lUP", "m4lDOWN"],
name="variation",
)
.Weight()
)
# ...
# fill histograms based on dataset_category
# ...
return {"data": mllllhist_data, "MC": mllllhist_MC}
def postprocess(self, accumulator):
pass
A coffea processor designed to make physics motivated event selections to create accumulators of the 4-lepton invariant mass.
3.3Feature engineering: The invariant mass¶
In order to discriminate the events of interest, i.e. candidates of the Higgs boson decay, from the vast background which has the same experimental signature, a discriminating feature is constructed. The example shown uses a simple, physics-inspired discriminant the “invariant mass” but the methods used can use complex feature engineering that involve machine learning methods to calculate more efficient discriminants. The invariant mass is the mass of a system that remains constant regardless of the system’s motion or the reference frame in which it is measured. Invariant mass is derived from the energy and momentum of a system of particles and is a fundamental property of the system:
where and is the total energy and momentum of the particles, respectively.
By detecting and measuring the energies and momenta of the detected particles at the experiment, we can reconstruct the invariant mass of the decay system. Particle systems originating from the decay of the Higgs boson will have a characteristic value of the invariant mass, which after the discovery in 2012 we know it is about . This is the quantity that will allow us to discriminate from particle systems that originate from background processes.
3.4Measurement uncertainties¶
One of the most expensive operations that happens during the event selections is the computation of systematic variations of the events to accommodate for imperfect knowledge of the detector systems.
This in practice requires applying complex, experiment specific corrections to each event, using algorithms implemented in C++.
Historically these tools were implemented for an event loop processing paradigm, but with recent tooling additions, as shown in Figure 1, efficient on-the-fly systematic corrections can be computed for array programming paradigms.
The following provides an example of high level Python APIs that provide handles to these tools to use in the workflows described so far. These tools are efficient enough to be able to apply multiple systematic variations in analysis workflows, as seen in Figure 2.
import awkward as ak
from atlascp import EgammaTools # ATLAS CP tool Python nanobind bindings
def get_corrected_mass(energyCorrectionTool, electrons, sys=None):
electron_vectors = ak.zip(
{
"pt": energyCorrectionTool(electrons, sys=sys).newPt,
"eta": electrons.eta,
"phi": electrons.phi,
"mass": electrons.m,
},
with_name="Momentum4D",
)
return (electron_vectors[:, 0] + electron_vectors[:, 1]).mass / 1000 # GeV
energy_correction_tool = EgammaTools.EgammaCalibrationAndSmearingTool()
# ...
# configure and initialize correction algorithm
# ...
energy_correction_tool.initialize()
corrected_m_Res_UP = get_corrected_mass(
energy_correction_tool, electrons, "Res_up"
).compute()
Simplified example of what the Python API for a systematic correction tool with a columnar implementation looks like.
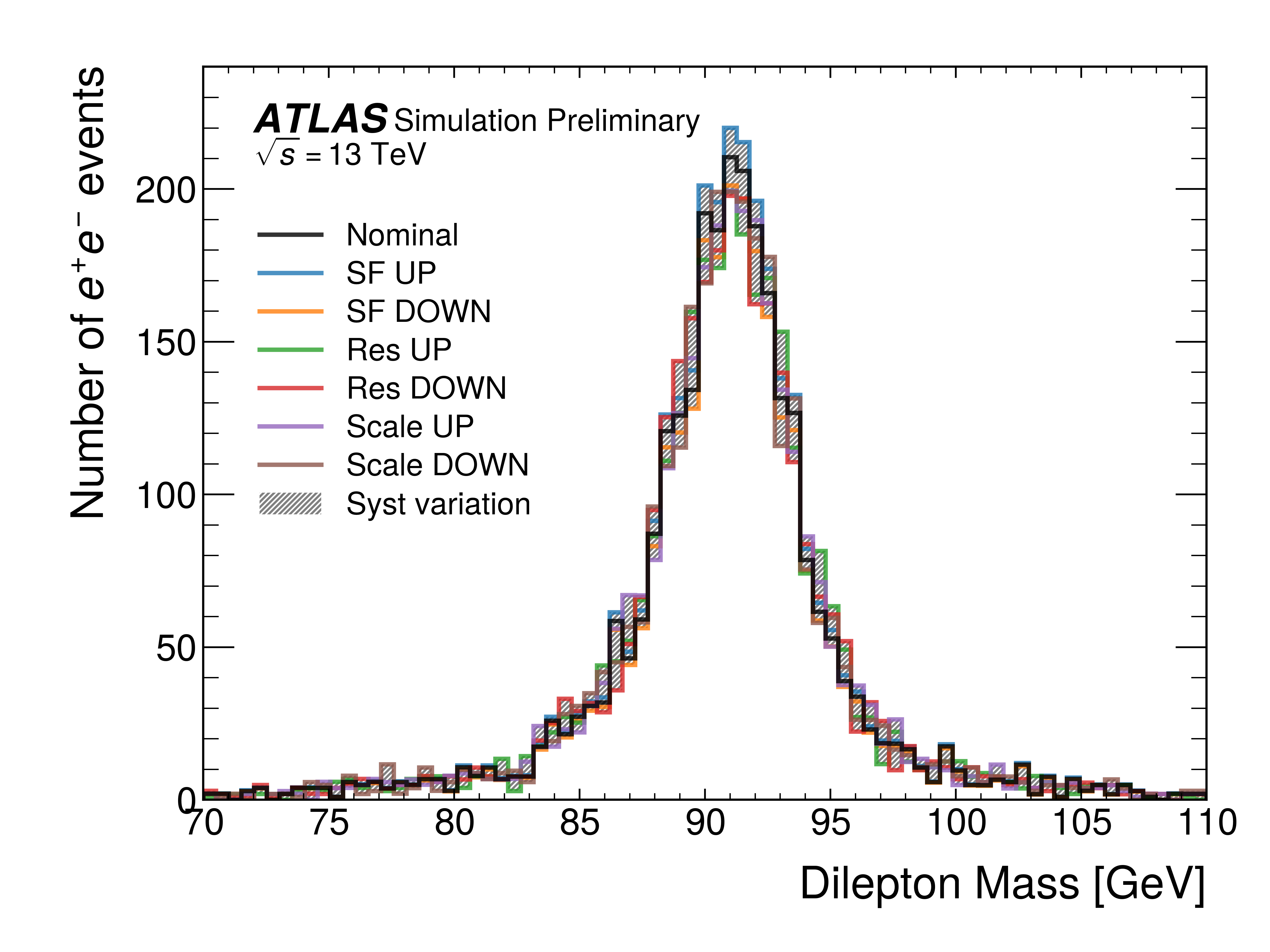
Figure 2:Example of the reconstructed dilepton invariant mass distribution in simulation with the electron reconstruction and identification efficiency scale factor (SF) and corrections to the energy resolution (res) energy scale (scale) computed on-the-fly using the nanobind Python bindings to the ATLAS C++ correction tools.
The total variation in the systematic corrections is plotted as a hashed band. Kourlitis et al., 2024
3.5The “discovery” plot¶
After running the coffea processors, the resulting data from the selections is accumulated into boost-histogram objects, as seen visualized in Figure 3.
import hist
import mplhep
mplhep.histplot(
all_histograms["data"], histtype="errorbar", color="black", label="Data"
)
hist.Hist.plot1d(
all_histograms["MC"][:, :, "nominal"],
stack=True,
histtype="fill",
color=["purple", "red", "lightblue"],
)
Using mplhep, hist, and matplotlib the post-processed histograms of the simulation and the data are visualized in advance of any statistical inference of best-fit model parameters.
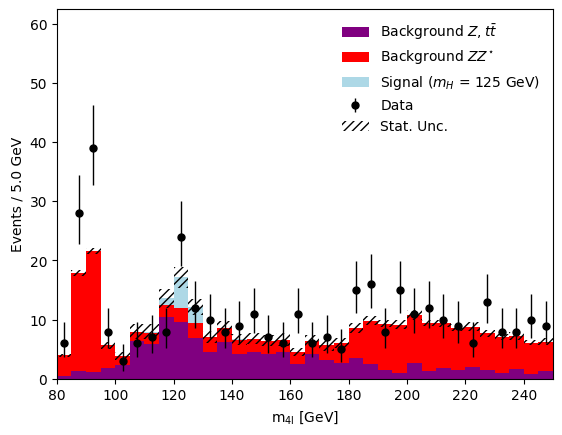
Figure 3:Using mplhep, hist, and matplotlib the post-processed histograms of the simulation and the data are visualized in advance of any statistical inference of best-fit model parameters.
These histograms are then serialized into files with uproot and used by the statistical modeling and inference libraries pyhf Heinrich et al., n.d.Heinrich et al., 2021 and cabinetry Held & Feickert, n.d. to build binned statistical models and efficiently fit the models to the observed data using vectorized computations and the optimization library iminuit Dembinski et al., n.d. for full uncertainties on all model parameters.
The resulting best-fit model parameters — such as the scale factor on the signal component of the model corresponding to the normalization on the Higgs contributions — are visualized in Figure 4, where good agreement between the model predictions and the data is observed.
The signal component, clearly visible above the additional “background” components of the model, are Higgs boson events, with an observed count in agreement with theoretical expectations.
import cabinetry
import numpy as np
config = cabinetry.configuration.load("config.yml")
cabinetry.templates.collect(config)
cabinetry.templates.postprocess(config) # optional post-processing (e.g. smoothing)
workspace = cabinetry.workspace.build(config)
model, data = cabinetry.model_utils.model_and_data(workspace)
fit_results = cabinetry.fit.fit(model, data)
# create post-fit model prediction
postfit_model = cabinetry.model_utils.prediction(model, fit_results=fit_results)
# binning to use in plot
plot_config = {
"Regions": [
{
"Name": "Signal_region",
"Binning": list(np.linspace(bin_edge_low, bin_edge_high, num_bins + 1)),
}
]
}
figure_dict = cabinetry.visualize.data_mc(
postfit_model, data, config=plot_config, save_figure=False
)
# modify x-axis label
fig = figure_dict[0]["figure"]
fig.axes[1].set_xlabel("m4l [GeV]")
Using cabinetry, pyhf, and matplotlib the data and the post-fit model prediction are visualized.
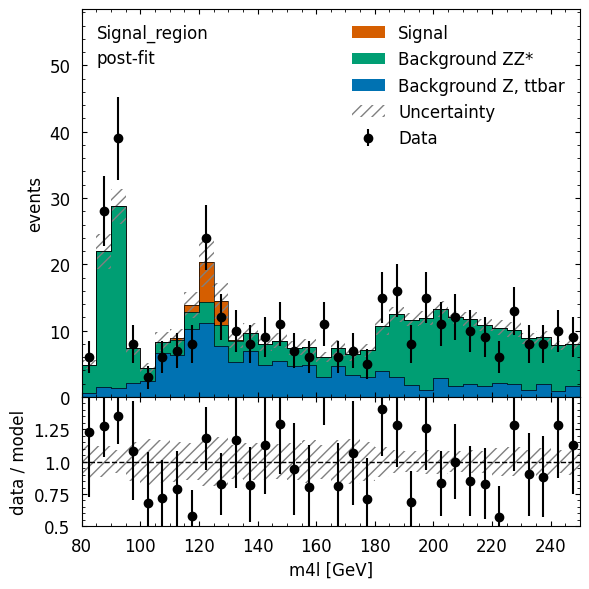
Figure 4:Using cabinetry, pyhf, and matplotlib the data and the post-fit model prediction are visualized.
4Conclusions¶
When the Higgs boson was discovered in 2012, the idea of being able to perform real Pythonic data analysis in HEP, let alone efficient analysis, was viewed as unfeasible. Though investment in the broader Scientific Python ecosystem, and development of the domain specific pieces in the Scikit-HEP organization the field of particle physics successfully created a PyHEP ecosystem of robust tooling. Further investment by the ATLAS collaboration has resulted in new performant tooling for complex systematic corrections that will allow for more full and complex operations to be performed entirely within a Python workflow, helping to further reduce the time to insight for physics analysts.
Matthew Feickert, Alexander Held, and Gordon Watts are supported by the U.S. National Science Foundation (NSF) under Cooperative Agreement OAC-1836650 and PHY-2323298 (IRIS-HEP). Lukas Heinrich and Vangelis Kourlitis are supported by the Excellence Cluster ORIGINS, which is funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy - EXC-2094-390783311 and by the German Federal Ministry of Education and Research Project 05H2021 (ErUM-FSP T02) - “Run 3 von ATLAS am LHC: Analysis Infrastructure for the ATLAS Detektor at the LHC”.
Copyright © 2024 Feickert et al. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license, which enables reusers to distribute, remix, adapt, and build upon the material in any medium or format, so long as attribution is given to the creators.
- CP
- Combined Performance
- EDM
- Event Data Model
- HEP
- High Energy Physics
- IRIS-HEP
- Institute for Research and Innovation in Software for High Energy Physics
- LHC
- Large Hadron Collider
- NSF
- National Science Foundation
- ATLAS Collaboration. (2008). The ATLAS Experiment at the CERN Large Hadron Collider. JINST, 3, S08003. 10.1088/1748-0221/3/08/S08003
- Rodrigues, E., & others. (2020). The Scikit HEP Project – overview and prospects. EPJ Web Conf., 245, 06028. 10.1051/epjconf/202024506028
- Henry Schreiner, Jim Pivarski, & Eduardo Rodrigues. (2022). Awkward Packaging: building Scikit-HEP. In Meghann Agarwal, Chris Calloway, Dillon Niederhut, & David Shupe (Eds.), Proceedings of the 21st Python in Science Conference (pp. 115–120). 10.25080/majora-212e5952-012
- Elmer, P., Neubauer, M., & Sokoloff, M. D. (2017). Strategic Plan for a Scientific Software Innovation Institute (S2I2) for High Energy Physics.
- Albrecht, J., & others. (2019). A Roadmap for HEP Software and Computing R&D for the 2020s. Comput. Softw. Big Sci., 3(1), 7. 10.1007/s41781-018-0018-8
- HEP Software Foundation. (2024). Python in HEP (PyHEP) HSF Working Group. https://hepsoftwarefoundation.org/workinggroups/pyhep.html
- Jakob, W. (2022). nanobind: tiny and efficient C++/Python bindings. https://github.com/wjakob/nanobind
- Harris, C. R., Millman, K. J., van der Walt, S. J., Gommers, R., Virtanen, P., Cournapeau, D., Wieser, E., Taylor, J., Berg, S., Smith, N. J., Kern, R., Picus, M., Hoyer, S., van Kerkwijk, M. H., Brett, M., Haldane, A., del Río, J. F., Wiebe, M., Peterson, P., … Oliphant, T. E. (2020). Array programming with NumPy. Nature, 585(7825), 357–362. https://doi.org/10.1038/s41586-020-2649-2
- Pivarski, J., Osborne, I., Ifrim, I., Schreiner, H., Hollands, A., Biswas, A., Das, P., Roy Choudhury, S., Smith, N., & Goyal, M. (2018). Awkward Array. 10.5281/zenodo.4341376
- Hartmann, N., Elmsheuser, J., & Duckeck, G. (2021). Columnar data analysis with ATLAS analysis formats. EPJ Web Conf., 251, 03001. 10.1051/epjconf/202125103001
- Brun, R., & Rademakers, F. (1997). ROOT: An object oriented data analysis framework. Nucl. Instrum. Meth. A, 389, 81–86. 10.1016/S0168-9002(97)00048-X
- Pivarski, J., Elmer, P., & Lange, D. (2020). Awkward Arrays in Python, C++, and Numba. EPJ Web Conf., 245, 05023. 10.1051/epjconf/202024505023
- Pivarski, J., Schreiner, H., Hollands, A., Das, P., Kothari, K., Roy, A., Ling, J., Smith, N., Burr, C., & Stark, G. (2017). Uproot. 10.5281/zenodo.4340632
- ATLAS Collaboration. (2021). The ATLAS Collaboration Software and Firmware. ATL-SOFT-PUB-2021-001. https://cds.cern.ch/record/2767187
- ATLAS Collaboration. (2021). Athena. 10.5281/zenodo.4772550
