Orchestrating Bioinformatics Workflows Across a Heterogeneous Toolset with Flyte
Abstract¶
While Python excels at prototyping and iterating quickly, it’s not always performant enough for whole-genome scale data processing. Flyte, an open-source Python-based workflow orchestrator, presents an excellent way to tie together the myriad tools required to run bioinformatics workflows. Flyte is a Kubernetes native orchestrator, meaning all dependencies are captured and versioned in container images. It also allows you to define custom types in Python representing genomic datasets, enabling a powerful way to enforce compatibility across tools. Finally, Flyte provides a number of different abstractions for wrapping these tools, enabling further standardization. Computational biologists, or any scientists processing data with a heterogeneous toolset, stand to benefit from a common orchestration layer that is opinionated yet flexible.
1Introduction¶
Since the sequencing of the human genome Venter et al., 2001, and as other wet lab processes have scaled in the last couple decades, computational approaches to understanding the living world have exploded. The firehose of data generated from all these experiments led to algorithms and heuristics developed in low-level high-performance languages such as C and C++. Later on, industry standard collections of tools like the Genome Analysis ToolKit (GATK) Auwera et al., 2013 were written in Java. A number of less performance intensive offerings such as MultiQC Ewels et al., 2016 are written in Python; and R is used extensively where it excels: visualization and statistical modeling. Finally, newer deep-learning models and Rust based components are entering the fray.
Different languages also come with different dependencies and approaches to dependency management, interpreted versus compiled languages for example handle this very differently. They also need to be installed correctly and available in the user’s PATH for execution. Moreover, compatibility between different tools in bioinformatics often falls back on standard file types expected in specific locations on a traditional filesystem. In practice this means searching through datafiles or indices available at a particular directory and expecting a specific naming convention or filetype.
In short, bioinformatics suffers from the same reproducibility crisis Baker, 2016 as the broader scientific landscape. Standardizing interfaces, as well as orchestrating and encapsulating these different tools in a flexible and future-proof way is of paramount importance on this unrelenting march towards larger and larger datasets.
2Methods¶
Solving these problems using Flyte is accomplished by capturing dependencies flexibly with dynamically generated container images, defining custom types to enforce at the task boundary, and wrapping tools in Flyte tasks. Before diving into the finer points, a brief primer on Flyte is advised. While the introduction in the docs is a worthwhile read before continuing, here is a more concise “hello world” example:
from flytekit import task, workflow
@task
def greet() -> str:
return "Hello"
@task
def say(greeting: str, name: str) -> str:
return f"{greeting}, {name}!"
@workflow
def hello_world_wf(name: str = "world") -> str:
greeting = greet()
res = say(greeting=greeting, name=name)
return resTasks are the most basic unit of work in Flyte. They are pure-Python functions and their interface is strongly typed in order to compose the workflow. The workflow itself is actually a domain-specific language that statically compiles a directed-acyclic graph (DAG) based on the dependencies between the different tasks. There are different flavors of tasks and workflows as we’ll see later, but this is the core concept.
The following example details a bioinformatics workflow built using Flyte. All code is drawn from the ever-evolving unionbio Github repository. There are many more datatypes, tasks and workflows defined there. Questions are always welcome and contributions are of course encouraged!
2.1Images¶
While it’s possible to run Flyte tasks and workflows locally in a Python virtual environment, production executions in Flyte run on a Kubernetes cluster. As a kubernetes native orchestrator, all tasks run in their own (typically single-container) pods using whatever image is specified in the @task decorator. Capturing dependencies in container images has been a standard for some time now, but this is taken a step further with ImageSpec. ImageSpec lets you easily define a base image and additional dependencies right alongside your task and workflow code. Additional dependencies from PyPI, Conda, or apt are supported out-of-box. Arbitrary RUN commands are also available for base images lacking Debian’s package manager, or to accomplish virtually anything not currently implemented. Finally, while envd is the default builder, other backends like a local Docker daemon or even remote builders are available should the need arise.
These ImageSpec definitions are loosely coupled to your workflow code and are built automatically when tasks are registered or run on a Flyte cluster. ImageSpec reduces the complexity inherent in manually authoring a Dockerfile and enables a more streamlined approach to building images without the need for an additional build step and configuration update to reference the latest image. This coupling and reduced complexity makes it easier to build single-purpose images instead of throwing everything into one monolithic image.
main_img = ImageSpec(
name="main",
platform="linux/amd64",
python_version="3.11",
packages=["flytekit"],
conda_channels=["bioconda"],
conda_packages=[
"samtools",
"bcftools",
"fastp",
"bowtie2",
"gatk4",
"fastqc",
],
registry="docker.io/unionbio",
)
folding_img = ImageSpec(
name="protein",
platform="linux/amd64",
python_version="3.11",
packages=["flytekit", "transformers", "torch"],
conda_channels=["bioconda", "conda-forge"],
conda_packages=[
"prodigal",
"biotite",
"bioPython",
"py3Dmol",
"matplotlib",
],
registry="docker.io/unionbio",
)The main image has a lot of functionality and could arguably be pared down. It contains a number of very common low-level tools, along with GATK and a couple aligners Armstrong et al., 2019. The protein image on the other hand, only contains a handful of tools related to a very specific protein folding and visualization workflow. Unless using a remote builder, these images are built locally and then pushed to the registry specified. They will persist in the builder’s local registry and leverage the builder’s cache until cleaned-up. Once built, they are Open Container Initiative (OCI) compliant container images like any other, allowing you to compose them as you see fit. The main image could be used as the base for the folding image, for example. Another very simple but powerful use case would be to Flytify any off-the-shelf image by simply specifying a Python version and adding flytekit as a package.
Currently, a subset of the full Dockerfile functionality has been reimplemented in ImageSpec. A typical Dockerfile could include pulling Micromamba binaries, creating and activating an environment, before finally installing the relevant packages. ImageSpec’s opinionated approach enables a simpler experience by handling this kind of boilerplate code behind-the-scenes. ImageSpec is also context aware in the same way docker build is, meaning a source_root containing a lock or env file can be specified and installed if you want to keep your local environment in sync. The images submodule of the unionbio repo puts this in practice, with different source roots used in test and production.
ImageSpecs can be specified in the task decorator alongside any infrastructure requirements in a very granular fashion:
@task(
container_image=folding_img,
requests=Resources(cpu="4", mem="32Gi", gpu="1"),
accelerator=GPUAccelerator("nvidia-tesla-v100"),
)
def predict_structure(seq: str):
fold_protein(seq)This image will be built and uploaded when your tasks and workflows are registered to a Flyte cluster.
2.2Datatypes¶
Having rich data types to enforce compatibility at the task boundary is essential to these wrapped tools working together. Flyte supports arbitrary data types through Python’s dataclasses library. Genomics pipelines typically pass around one or many large text files related to the same sample. Data types capturing these files allow us to reason about them and their metadata more easily across tasks, as well as enforce naming conventions.
Importantly, Flyte abstracts the object store, allowing you to load these assets into pods wherever is most convenient for your tool. This not only makes it easier to work with these files, but also safer as you’re working with ephemeral storage during execution instead of a shared production filesystem. In a shared filesystem, unintended side-effects could mutate artifacts unrelated to the current production run. This can be mitigated in a number of ways, such as setting up an empty directory for every experiment or restricting permissions to files after a run is complete. In an ephemeral setting however, inputs of interest are materialized at the beginning of the task and any relevant outputs are serialized to a unique prefix in the object store when the task completes. Any unintended modifications disappear when the pod is deleted.
Sequencers employ different strategies to produce millions of short strings representing DNA fragments called reads. One such strategy is single-read sequencing, which produces one sequencing read per DNA fragment. This is appropriate for simpler analyses such as profiling or for analyzing less complex genomes. Paired-end sequencing, on the other hand, produces two reads representing both sequencing directions of the DNA fragment. This is essential in applications such as “de novo” assembly which involves re-assembling these reads into a whole genome without a known reference as a guide. It also unlocks the detection of larger variations, typically longer insertions or deletions.
Regardless of the sequencing strategy, these reads are captured in one or a pair of FastQ files, an example of which is given below:
@SRR001666.1 071112_SLXA-EAS1_s_7:5:1:817:345 length=72
GGGTGATGGCCGCTGCCGATGGCGTCAAATCCCACCAAGTTACCCTTAACAACTTAAGGGTTTTCAAATAGA
+SRR001666.1 071112_SLXA-EAS1_s_7:5:1:817:345 length=72
IIIIIIIIIIIIIIIIIIIIIIIIIIIIII9IG9ICIIIIIIIIIIIIIIIIIIIIDIIIIIII>IIIIII/
@SRR001666.2 071112_SLXA-EAS1_s_7:5:1:801:338 length=72
GTTCAGGGATACGACGTTTGTATTTTAAGAATCTGAAGCAGAAGTCGATGATAATACGCGTCGTTTTATCAT
+SRR001666.2 071112_SLXA-EAS1_s_7:5:1:801:338 length=72
IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII6IBIIIIIIIIIIIIIIIIIIIIIIIGII>IIIII-I)8ITwo reads are present here, characterized by the nucleobases A, C, T, and G. The other lines contain metadata and quality information. Here is the dataclass that encapsulates these files and any salient information about them:
@dataclass
class Reads(DataClassJSONMixin):
sample: str
filtered: bool | None = None
filt_report: FlyteFile | None = None
uread: FlyteFile | None = None
read1: FlyteFile | None = None
read2: FlyteFile | None = None
def get_read_fnames(self):
filt = "filt." if self.filtered else ""
return (
f"{self.sample}_1.{filt}fastq.gz",
f"{self.sample}_2.{filt}fastq.gz",
)
def get_report_fname(self):
return f"{self.sample}_fastq-filter-report.json"
@classmethod
def make_all(cls, dir: Path):
...We’re capturing a few important aspects: whether the reads have been filtered and the results of that operation, as well as if they’re paired-end reads or not. Paired-end reads will populate the read1 and read2 attributes. If they are unpaired then a single FastQ file representing a sample’s reads is defined in the uread field. The presence or absence of these attributes implicitly disambiguates the sequencing strategy.
Additionally, the make_all function body has been omitted for brevity, but it accepts a directory and returns a list of these objects based on it’s contents. In the other direction, a get_read_fnames method is defined to standardize naming conventions based on The 1000 Genomes Project guidelines.
FlyteFile, along with FlyteDirectory, represent a file or directory in a Flyte aware context. These types handle serialization and deserialization into and out of the object store. They re-implement a number of common filesystem operations like open(), which returns a streaming handle, for example. Simply returning a FlyteFile from a task will automatically upload it to whatever object store is defined. This unassuming piece of functionality is one of Flyte’s key strengths: abstracting data management so researchers can focus on their task code. Since dataflow in Flyte is a first-class construct, having well defined inputs and outputs at the task boundary makes authoring workflows that much more reliable.
In order to accomplish sequencing in a sensible timeframe, reads generation is massively parallelized Venter et al., 1998. This dramatically improves the throughput, but removes crucial information regarding the location of those reads. In order to recover that information, the reads are aligned to a known reference genome, producing an Alignment file, which we also capture in a dataclass:
@dataclass
class Alignment(DataClassJSONMixin):
sample: str
aligner: str
format: str | None = None
alignment: FlyteFile | None = None
alignment_idx: FlyteFile | None = None
alignment_report: FlyteFile | None = None
sorted: bool | None = None
deduped: bool | None = None
bqsr_report: FlyteFile | None = None
def _get_state_str(self):
state = f"{self.sample}_{self.aligner}"
if self.sorted:
state += "_sorted"
if self.deduped:
state += "_deduped"
return state
def get_alignment_fname(self):
return f"{self._get_state_str()}_aligned.{self.format}"
@classmethod
def make_all(cls, dir: Path):
...Compared to the Reads dataclass, the attributes captured here are of course only relevant to Alignments. However, the methods that interact with the local filesystem and enforce naming conventions remain. In the next section, we’ll look at tasks that actually carry out this alignment.
2.3Tasks¶
While Flyte tasks are written in Python, there are a few ways to wrap arbitrary tools. ShellTasks are one such way, allowing you to define scripts as multi-line strings in Python. For added flexibility around packing and unpacking data types before and after execution, Flyte also ships with a subproc_execute function which can be used in vanilla Python tasks. Finally, arbitrary images can be used via a ContainerTask and avoid any flytekit dependency altogether.
Bowtie2 Langmead et al., 2009, a fast and memory efficient aligner, is used to carry out the aforementioned alignments. Before alignment can be carried out efficiently, an index must be generated from the reference genome. Indices are generated by pre-processing the reference into a data structure that enables rapid lookup of a match to a given read. Bowtie2 uses an FM-index which combines the Burrows-Wheeler Transform (BWT) with a suffix array. Broadly speaking, BWT enables compression of the data while the suffix array allows for efficient lookup of substrings. Here is a ShellTask creating a bowtie2 index directory from a genome reference file:
bowtie2_index = ShellTask(
name="bowtie2-index",
debug=True,
requests=Resources(cpu="4", mem="10Gi"),
metadata=TaskMetadata(retries=3, cache=True, cache_version=ref_hash),
container_image=main_img,
script="""
mkdir {outputs.idx}
bowtie2-build {inputs.ref} {outputs.idx}/bt2_idx
""",
inputs=kwtypes(ref=FlyteFile),
output_locs=[
OutputLocation(var="idx", var_type=FlyteDirectory, location="/tmp/bt2_idx")
],
)This task uses the main_img defined above; it also accepts a FlyteFile and outputs a FlyteDirectory. Another important feature to highlight here is caching, which saves us valuable compute for inputs that rarely change. Since the alignment index for a particular aligner only needs to be generated once for a given reference, we’ve set the cache_version to a hash of the reference’s URI. As long as the reference exists at that URI, this bowtie indexing task will complete immediately and return that index. To perform the actual alignment, a regular Python task is used with a Flyte-aware subprocess function to call the bowtie CLI.
@task(container_image=main_img, requests=Resources(cpu="4", mem="10Gi"))
def bowtie2_align_paired_reads(idx: FlyteDirectory, fs: Reads) -> Alignment:
idx.download()
ldir = Path(current_context().working_directory)
alignment = Alignment(fs.sample, "bowtie2", "sam", sorted=False, deduped=False)
al = ldir.joinpath(alignment.get_alignment_fname())
rep = ldir.joinpath(alignment.get_report_fname())
cmd = [
"bowtie2",
"-x",
f"{idx.path}/bt2_idx",
"-1",
fs.read1.path,
"-2",
fs.read2.path,
"-S",
al,
]
result = subproc_execute(cmd)
# Bowtie2 alignment writes stats to stderr
with open(rep, "w") as f:
f.write(result.error)
alignment.alignment = FlyteFile(path=str(al))
alignment.alignment_report = FlyteFile(path=str(rep))
return alignmentSince Python tasks are the default task type, they’re the most feature rich and stable. The main advantage to using one here is to unpack the inputs and construct the output type.
The resulting alignments are then deduplicated, sorted, recalibrated and reformatted. While important, these tasks are similarly implemented to the above alignment function and as such are omitted to focus on the next major step: variant calling. Variant calling has such a diversity of approaches that any meaningful exploration of the landscape is out of scope. However, the central purpose is to distill the aligned reads into a set of likely relevant loci that deviate from the reference. This is accomplished by aggregating information like quality scores and the number of reads covering a given location, called a pileup, to come to consensus around a most likely call.
@task(container_image=main_img_fqn)
def haplotype_caller(ref: Reference, al: Alignment) -> VCF:
ref.aggregate()
al.aggregate()
vcf_out = VCF(sample=al.sample, caller="gatk-hc")
vcf_fn = vcf_out.get_vcf_fname()
vcf_idx_fn = vcf_out.get_vcf_idx_fname()
hc_cmd = [
"gatk",
"HaplotypeCaller",
"-R",
ref.get_ref_path(),
"-I",
al.alignment.path,
"-O",
vcf_fn,
]
subproc_execute(hc_cmd)
vcf_out.vcf = FlyteFile(path=vcf_fn)
vcf_out.vcf_idx = FlyteFile(path=vcf_idx_fn)
return vcf_outGATK’s HaplotypeCaller is used to perform variant calling by accepting an Alignment and Reference object and producing a Variant Call Format (VCF) file. VCFs are another common tabular text file, with each row representing a variant present in the input sequences and its alternate in the reference, along with a quality score and extensible columns for adding additional information. Variants can range from single-nucleotide polymorphisms (SNPs) to much larger structural variations mentioned above. Once variants are called, they can be further filtered downstream for certain characteristics, or against one of the many curated databases, to arrive at a set of actionable insights.
3Results¶
A real world variant discovery workflow demonstrates how to tie all these disparate parts together. Starting with a directory containing raw FastQ files, we’ll perform quality-control (QC), filtering, index generation, alignment, calling, and conclude with a final report of all the steps. Here’s the code:
from flytekit import workflow, dynamic, map_task
from flytekit.types.directory import FlyteDirectory
from flytekit.types.file import FlyteFile
from unionbio.tasks.utils import prepare_raw_samples
from unionbio.tasks.fastqc import fastqc
from unionbio.tasks.fastp import pyfastp
from unionbio.tasks.bowtie2 import bowtie2_idx, bowtie2_align_samples
from unionbio.tasks.callers import hc_call_variants
from unionbio.tasks.multiqc import render_multiqc
@workflow
def variant_discovery_wf(seq_dir: FlyteDirectory, ref_path: FlyteFile) -> FlyteFile:
# Generate FastQC reports and check for failures
fqc_out = fastqc(seq_dir=seq_dir)
samples = prepare_raw_samples(seq_dir=seq_dir)
# Map out filtering across all samples and generate indices
filtered_samples = map_task(pyfastp)(rs=samples)
# Explicitly define task dependencies
fqc_out >> filtered_samples
# Generate a bowtie2 index or load it from cache
bowtie2_idx = bowtie2_index(ref=ref_path)
# Generate alignments using bowtie2
sams = bowtie2_align_samples(idx=bowtie2_idx, samples=filtered_samples)
# Call variants
vcfs = hc_call_variants(ref=ref_path, als=sams)
# Generate final multiqc report with stats from all steps
return render_multiqc(fqc=fqc_out, filt_reps=filtered_samples, sams=sams, vcfs=vcfs)To help make sense of the flow of tasks, here is a screenshot from the Flyte UI that offers a visual representation of the different steps:
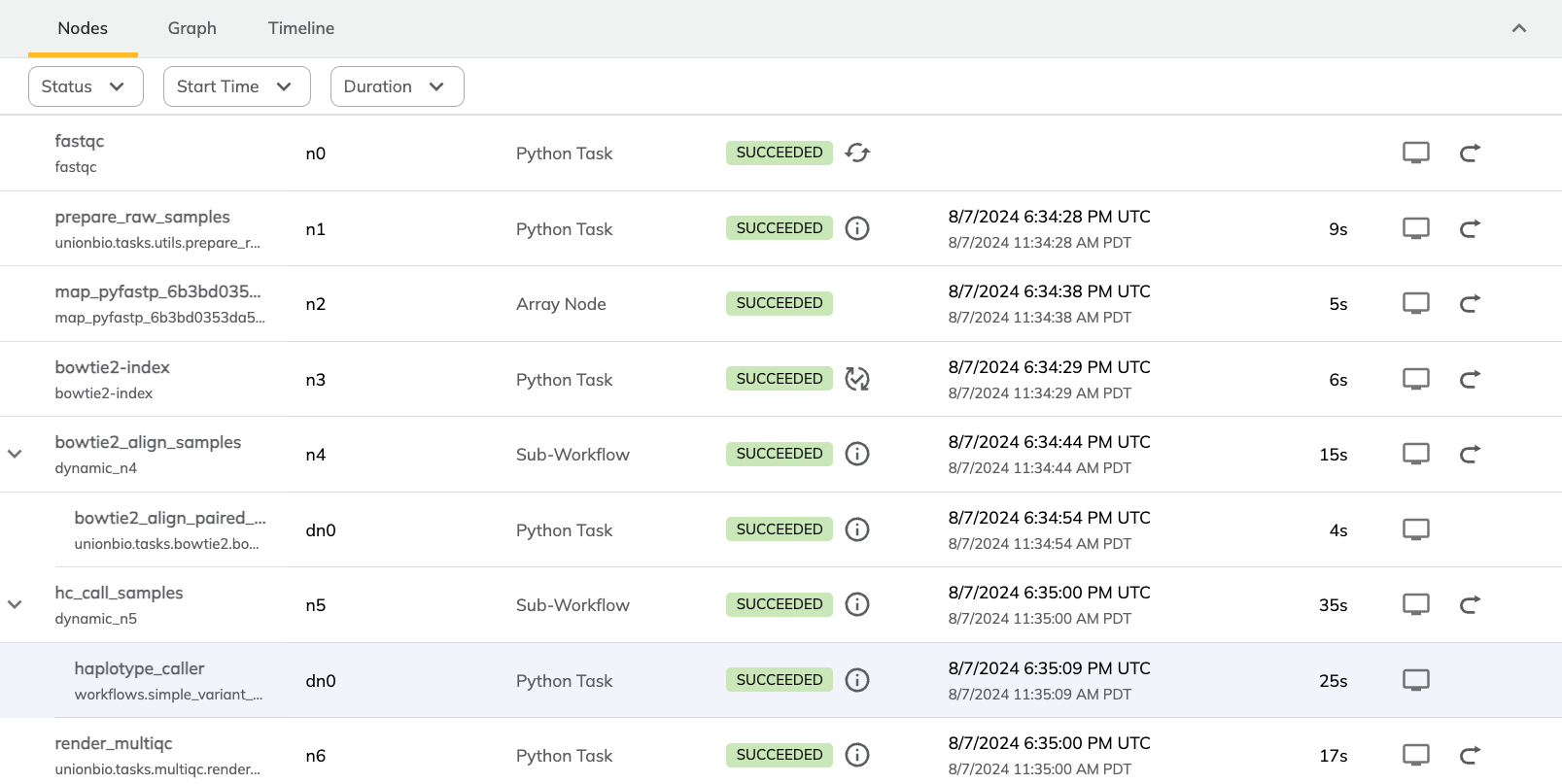
Figure 1:Table listing the various tasks of the workflow alongside task type, status, completion time, and runtime
FastQC Andrews et al., 2012, an extremely common QC tool, is wrapped in a ShellTask and starts off the workflow by generating a report for all FastQ formatted reads files in a given directory. That directory is then turned into Reads objects via the prepare_raw_samples task. Those samples are passed to fastp for adapter removal and filtering of duplicate or low quality reads. FastP Chen, 2023 is wrapped in a Python task which accepts a single Reads object. This task is then used in a map task to parallelize the processing of however many discrete samples were present in the input directory. Flyte relies on the flow of strongly-typed inputs and outputs to assemble the workflow; since there is no implicit dependency between filtering and QC, we make this relationship explicit with the “>>” operator.
Once pre-processing is complete, alignment can take place. First, bowtie2_index generates an index if one is not already cached. Since the bowtie2 alignment task processes samples one at a time, it was wrapped in a dynamic workflow to process a list of inputs. Dynamics are another parallelism construct, similar to map tasks with some key differences: they are more flexible than map tasks at the expense of some efficiency. Similarly, variant calling is performed across all samples using HaplotypeCaller. Lastly, MultiQC Ewels et al., 2016, produces a final report of all the different steps in the workflow. Certain task definitions are omitted for the sake of cogency, they are all fully-defined in the unionbio repo.
Despite being a fairly parsimonious workflow, it’s important to highlight how many different languages are seamlessly integrated. The preprocessing tools are written in Java and C/C++. Alignment is carried out with a mix of Perl and C++. HaplotypeCaller is implemented purely in Java. Finally, the reporting tool is implemented in Python. While this simplicity affords easy understanding of task-flow from the code, the Flyte console provides excellent visualizations to best understand it’s structure:
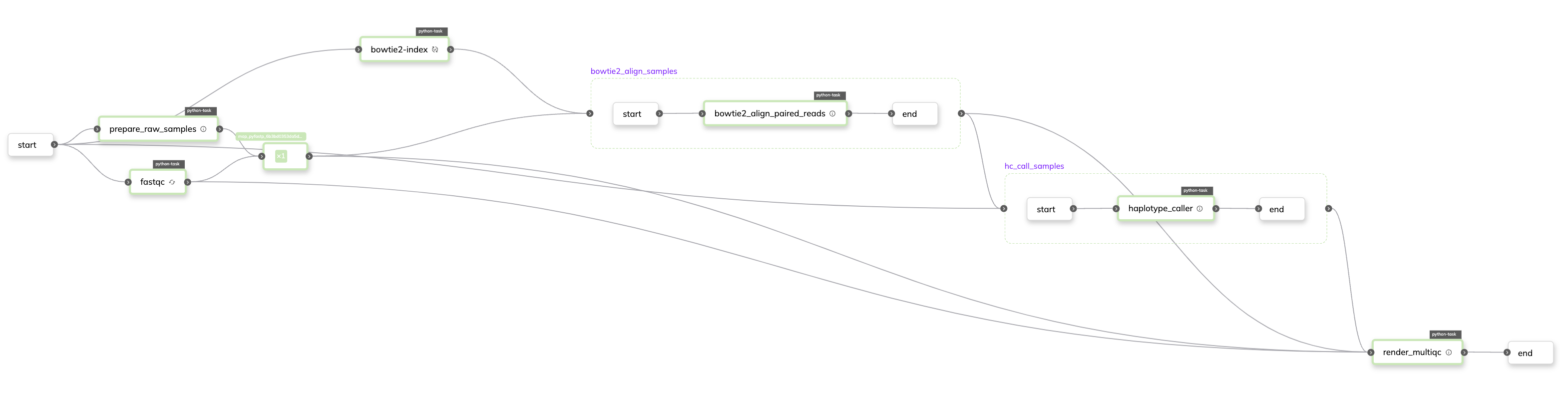
Figure 2:Workflow DAG showing the tasks as color-coded nodes with connections between them representing dependencies
Finally, it’s helpful to inspect a timeline of the execution which highlights a few things. Since this workflow was run several times over the course of capturing these figures, the fastqc task was cached in previous runs. It’s also clear from this figure which tasks were run in parallel in contrast to those which had dependencies on upstream outputs. Finally, the overall runtime is broken down into it’s separate parts. Since this was run on test data, everything executed fairly quickly.
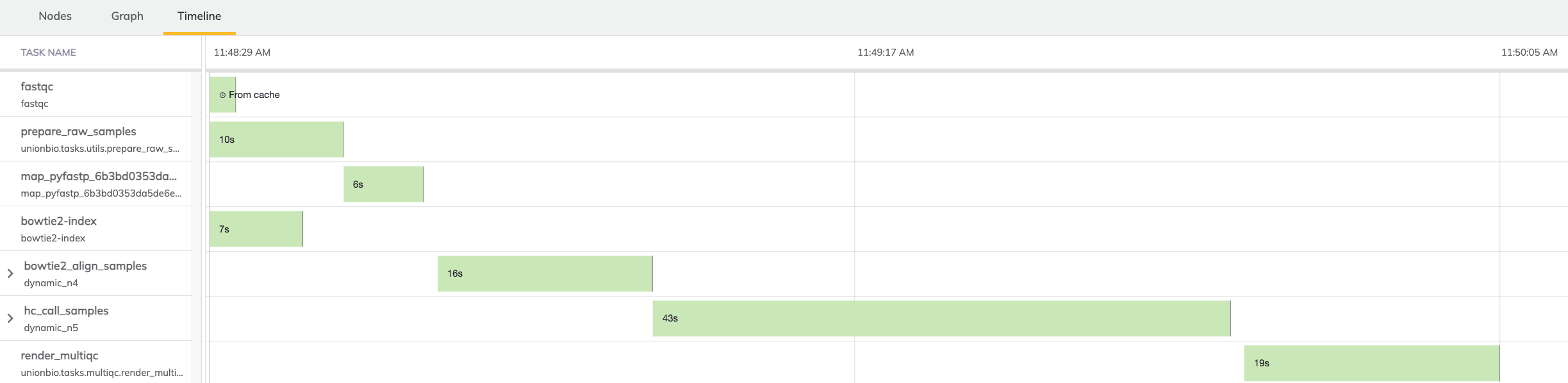
Figure 3:Execution timeline listing individual task runtimes in context of overall workflow runtime
4Conclusion¶
Different steps in a bioinformatics pipeline often require tools with significantly different characteristics. As such, different languages are employed where their strengths are best leveraged. Regardless of which language or framework is used, ImageSpec captures those dependencies in an OCI-compliant image for use in Flyte workflows and beyond in a very ergonomic way. Defining dataclasses with FlyteFiles and additional metadata frees the data flow from the trappings of a traditional filesystem, while Flyte handles serialization so we can easily operate in a cloud native paradigm. With dependencies handled in a robust way and the data interface standardized, wrapping arbitrary tools in Flyte tasks produces reusable and composable components that behave predictably. Tying all of this into a common orchestration layer presents an enormous benefit to the developer experience and consequently the reproducibility and extensibility of the research project as a whole.
- Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., Gocayne, J. D., Amanatides, P., Ballew, R. M., Huson, D. H., Wortman, J. R., Zhang, Q., Kodira, C. D., Zheng, X. H., Chen, L., … Zhu, X. (2001). The Sequence of the Human Genome. Science, 291(5507), 1304–1351. 10.1126/science.1058040
- Van der Auwera, G. A., Carneiro, M. O., Hartl, C., Poplin, R., del Angel, G., Levy‐Moonshine, A., Jordan, T., Shakir, K., Roazen, D., Thibault, J., Banks, E., Garimella, K. V., Altshuler, D., Gabriel, S., & DePristo, M. A. (2013). From FastQ Data to High‐Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Current Protocols in Bioinformatics, 43(1). 10.1002/0471250953.bi1110s43
- Ewels, P., Magnusson, M., Lundin, S., & Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics, 32(19), 3047–3048. 10.1093/bioinformatics/btw354
- Baker, M. (2016). 1,500 scientists lift the lid on reproducibility. Nature, 533(7604), 452–454. 10.1038/533452a
- Armstrong, J., Fiddes, I. T., Diekhans, M., & Paten, B. (2019). Whole-Genome Alignment and Comparative Annotation. Annual Review of Animal Biosciences, 7(1), 41–64. 10.1146/annurev-animal-020518-115005
- Venter, J. C., Adams, M. D., Sutton, G. G., Kerlavage, A. R., Smith, H. O., & Hunkapiller, M. (1998). Shotgun Sequencing of the Human Genome. Science, 280(5369), 1540–1542. 10.1126/science.280.5369.1540
- Langmead, B., Trapnell, C., Pop, M., & Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology, 10(3). 10.1186/gb-2009-10-3-r25
- Andrews, S., Krueger, F., Segonds-Pichon, A., Biggins, L., Krueger, C., & Wingett, S. (2012). FastQC. Babraham Institute.
- Chen, S. (2023). Ultrafast one‐pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta, 2(2). 10.1002/imt2.107
