
Continuous Tools for Scientific Publishing
Abstract¶
Advances in technology for data workflows have increased the speed and scope of scientific discovery, however, scientific dialogue still uses outdated technology for communicating and sharing knowledge. The widespread reliance on static PDF formats for research papers starkly contrasts with the complex, data-driven and increasingly computational nature of modern science. This gap, which is especially evident in the computational sciences, impedes the speed of research dissemination, reuse, and uptake. We require new mediums to compose ideas and ways to share research findings iteratively, as early as possible and connected directly to software and data. In this paper we discuss two tools for scientific authoring and publishing, MyST Markdown and Curvenote, and illustrate examples of improving metadata, reimagining the reading experience, including computational content, and transforming publishing practices for individuals and societies through automation and continuous practices. We focus on the unique aspects of the tools, which enable computational and interactive content, publishing and sharing continuously through automated checking and typesetting, and provide case studies from individuals to societies who have adopted these tools.
1Introduction and Motivation¶
In the face of mounting global challenges such as climate change, pandemics, and water security, the imperative for rapid, effective scientific discovery and dissemination has never been more acute. The pace at which these problems evolve and impact societies worldwide demands an equally dynamic and innovative approach to how scientific research is published. Despite significant advancements in technologies that enhance data collection, analysis, and workflow efficiency, the mechanisms through which scientific knowledge is shared have remained largely unchanged for decades Bourne et al., 2012 [1]. The widespread reliance on static PDF formats for research papers starkly contrasts with the complex, data-driven and increasingly computational nature of modern science, creating bottlenecks in knowledge dissemination and uptake.
This paper documents some of the design decisions made to address challenges in science communication and publishing in two tools:
(1) MyST Markdown (Markedly Structured Text, https://mystmd.org), a community-run open-source Jupyter sub-project[2], which is a text-based authoring framework that integrates computational content (e.g. Jupyter Notebooks); and
(2) Curvenote (https://
In developing these integrated tools and workflows, our goal is to lower the barriers to continuously releasing and iterating on scientific ideas in the open and address the related challenges of authoring and publishing in the context of computational, open-science documents. Introducing authoring tools that can understand and express structured, interactive, and computational content has the potential to fundamentally change the way scientific writing is checked, shared, and published — enabling faster iterations and direct ties to reproducible, interactive content.
1.1Authoring Structured Content¶
There are currently many challenges for individuals or groups to author research information that can be shared in a structured and rigorous way. By this we mean the things that structurally set a scientific article apart from, for example, a blog post: structured content, cross-references, valid citations with persistent identifiers (PIDs), and standardized metadata for licensing, funding information, authors, and affiliations. These structured content and metadata, as well as the standards behind them, are what define the “scientific record” and enable archiving, discoverability, accessibility, interoperability and the ability to reuse or cite content Wilkinson et al., 2016. One metric for measuring the difficulty of satisfying these scientific standards is to look at the direct costs that are spent on transforming author submissions (e.g. a PDF or a Word Document) into something that conforms to these standards and is ultimately archived. In scientific publishing, about 15% of Article Processing Charges (APCs) go to direct publication costs[4] Grossmann & Brembs, 2021. When applied to the global publishing industry[5], this suggests that approximately USD$2 billion dollars is spent on transforming author submissions (e.g. a word-document, LaTeX, or a PDF) into a copyedited, well-formatted, typeset document that can be archived with appropriate metadata Hagve, 2020. This estimate does not include the approximately USD$230 million spent on reformatting articles by scientists before publication Clotworthy et al., 2023. Many of these processes are hidden from authors[6] as well as actionable access to many of the benefits of structured data beyond citation graphs.
One goal of the MyST Markdown project is to dramatically reduce these direct-publication costs[7] and provide unfettered access to structured data as an output of authoring. The availability of this structured data directly enables exported content in a variety of formats including HTML, PDF and JATS-XML (a NISO standard for archiving scientific articles). In this article, we will demonstrate that having structured data throughout authoring can lead to a number of novel reading and authoring experiences Example 1, connect to interactivity and computation Example 2, and can provide new opportunities for reuse and quality checks when publishing Example 3. Furthermore, these transformation processes can be run continuously, opening the possibilities for faster feedback see Section 1.3, iterative drafts, small tweaks and versioned improvements that otherwise would not be worth the time and cost.
1.2Computational Articles¶
A compounding challenge to scientific publishing that we are exploring through MyST Markdown and Curvenote is how to deeply integrate computational workflows and content into science communication to promote interactive explorations and reproducible practices. There are a host of challenges from user-interface design, to maintenance, to archiving computational content. Many other tools have worked on aspects of integrating computation into scientific articles, notably R-Markdown Xie et al., 2018 and it’s successor Quarto (https://quarto.org); both of these projects have similar aims to MyST Markdown. From a user-experience goals perspective, we are interested in questions such as:
- how to make a change in a notebook figure and have that immediately show up in a document;
- how to ensure computed values are inserted directly from source, rather than through copy-and-paste;
- how to expose interactivity and exploration that a researcher often has when analyzing a data-set;
- how to provide and launch archived interactive computing environments.
These questions require authoring tools to be able to execute content (e.g. using MyBinder; Project Jupyter et al., 2018), to integrate and display computational/interactive outputs directly in reading experiences, as well as scientific publishing systems that can understand and archive computational content (e.g. Docker containers). This deep integration can open up possibilities of embedding interactive visualizations and computational notebooks directly into scientific documents e.g. Example 2, transforming articles from static texts into rich, interactive, reproducible narratives. In 2023, the authors helped to lead several working groups related to these challenges as part of Notebooks Now!, a Sloan Foundation funded project led by the American Geophysical Union. Those working groups found that integrating computational documents, via Jupyter Notebooks, into scholarly publishing system requires a re-imagination of the publishing processes (from submission to peer-review to production to reading) and that many existing processes and platforms are ill-equipped to handle computational articles see Caprarelli et al., 2023. The “executable research articles” project out of eLife Tsang & Maciocci, 2020 has similar aims to Notebooks Now!, with some differences in how notebooks and articles are separated which we will discuss in Section 2.3.
The ability to deeply link computational content into how we communicate science can improve reproducible practices, and surface more interlinked content about methods and algorithms in use. If used to their full extent, these can also fully integrate live computational environments into scientific articles, which provides many exciting possibilities for interrogating and extending scientific data and methods Heidt, 2024.
1.3Continuous Science Practices¶
The manual effort involved in article production Section 1.1 coupled with the inability to integrate computational work Section 1.2 negatively impacts the number of iterations/versions and the immediacy of feedback to authors[8]. In other disciplines, such as software development, these metrics of iteration and rapid feedback are often highly encouraged, measured and constantly improved Mishra & Otaiwi, 2020Leite et al., 2019Chen, 2015. For example, software organizations often measure and improve: the release cadence of a software product (e.g. continuous delivery); how confident you are in that release (e.g. based on continuous integration tests); how you get early feedback and confidence from linters and tests (e.g. the speed of your unit tests and integrated linters into development environments); and how fast you can obtain feedback from real usage and users on in-progress work (e.g. observability, analytics, customer interviews, design prototypes). Continuous delivery practices of software development are also extremely well studied, with large-scale surveys of organizational performance, design, robustness, and speed (see Leite et al. (2019) and references within). One industry survey based on 36,000 professionals worldwide grouped and compared respondents based on software delivery performance DORA, 2018. The highest performing teams were 46x faster to release to production than the lowest performing teams (i.e. on-demand and multiple times per day vs monthly or bi-annually) and had 7x fewer errors (due in part to better continuous deployment and testing infrastructure as well as smaller changesets)[9] DORA, 2018. Similarly, Blinde (2022) found in a survey of 123 professionals that “deployment frequency” correlates the strongest with all other organizational performance metrics. The study concluded that “practices that improve lead time” (automated deployments, continuous testing and version control) have a positive impact on “both software delivery performance and organizational performance” Blinde, 2022. In a continuous deployment transformation over a year, Callanan & Spillane (2016) saw a 20x reduction in manual effort releasing software and a 7x speed up in releases to production.
The analogy between scientific publishing and software releases is imperfect and non-prescriptive (i.e. scientific research is very different than developing a product). However, the analogy is illustrative in areas where there is a focus on iterations, smaller changesets and releasing in-progress work[10] as soon as possible to get feedback from peers (i.e. scientific peer-review) or users (in the case of software products). The speed of scientific progress depends in part on the speed of iteration and feedback. The time it takes for the peer-review process is over three months, in high profile journals like Nature that time has almost doubled over the past decade Powell, 2016. Rejections are anywhere from 50-90% Grossmann & Brembs, 2021 with valuable reviews and expertise coming months or even years after the work is completed[11]. There are wide spread efforts in scientific publishing that focus on sharing smaller components of research (e.g. FigShare Jones & Hahnel, 2016, Octopus Martinez Garcia et al., 2023, MicroPublications Clark et al., 2014, NanoPublications Groth et al., 2010, Protocols Teytelman et al., 2016, PreRegistrations Nosek et al., 2018), automated tools in the publication process Smith et al., 2018, as well as sharing research sooner in the life cycle especially through preprints Lenharo, 2024, “Preprints in Progress” Penfold & Polka, 2020 and getting feedback sooner from a more diverse community e.g. PREreview Johansson & Saderi, 2020.
We refer to these related concepts as “continuous science”, adopting language and concepts from “continuous integration and deployment”. The mechanisms to support continuous processes are through automation, rapid feedback on errors, and focusing on small, rapid changesets to accelerate feedback from peers. This gives us a technical lens to assess, for example:
- How long does it take to get feedback if your metadata or DOI is incorrect?
- When a computational figure or data output changes, how long does it take to integrate that into your document?
- How can you assess and test that the structure of a document applies to editorial rules?
In science there is a highly manual, absurdly expensive and disconnected process between authoring and publishing. By moving to continuous practices and investing in the appropriate infrastructure to support continuous science we believe there is an opportunity to accelerate scientific discovery by multiple orders of magnitude while simultaneously increasing reproducibility, robustness and transparency of the underlying science.
1.4Article Outline¶
For research-communication to be transformative on a similar scale as continuous practices for software delivery, researchers require modern tools for authoring and publishing. There are two inter-related capabilities that are necessary for this transition:
- authoring mediums that support data, computation and structured content without the need for expensive typesetting; and
- publishing that is open and accessible to researchers at a variety of scales – individual publishing, lab-groups, societies and institutions.
Through the lens of MyST Markdown and Curvenote, this paper will explore how these tools aim to address critical gaps in current scientific publishing practices. Our motivation is to enhance the speed and impact of research dissemination, fostering a scientific ecosystem that is more collaborative, reproducible, and equipped to tackle the urgent global challenges of our time.
2Authoring Tools¶
MyST Markdown (Markedly Structured Text, https://mystmd.org) is a community-driven markup language that is a superset of CommonMark (a standard form of Markdown) with special syntax for citations, cross-references, and block and inline extension points called “directives” and “roles”.
The block-level content provides multi-line containers surrounded by either backticks or colons; examples include callout panels, figures, equations and tables (see documentation).
There is also specialized support for the types of metadata that are important to collect for scientific articles (funding, ORCIDs, CRediT Roles, etc.).
In 2022, the Executable Books project (https://mystmd command line interface (CLI), which was initially developed as the Curvenote CLI, and later transferred to the ExecutableBooks project.
In June 2024, MyST Markdown officially became part of Project Jupyter (see enhancement proposal).
This tool allows authors writing in MyST Markdown to easily build websites and documents and supports the JupyterLab MyST plugin.
MyST is influenced by reStructuredText (RST) and Sphinx – pulling on the nomenclature and introducing additional standards where appropriate.
There are also many intentional syntax similarities of MyST Markdown to R-Markdown Xie et al., 2018, Pandoc, and Quarto (https://quarto.org), especially in citation syntax and frontmatter structure.
MyST Markdown is written in Javascript and builds upon the unified community of tools, output formats and transforms, whereas Quarto builds upon Pandoc written in Haskell + Lua; there are also differences in approach in the legal and community foundations (Pandoc and Quarto are GPL vs. MyST which is a more permissive MIT license; MyST is a community-driven project by Project Jupyter whereas Quarto is driven by a single corporate entity).
The initial use case driving the development and design of MyST Markdown has been JupyterBook, which can create educational online textbooks and tutorials with Jupyter Notebooks and narrative content written in MyST.
This article will not attempt to describe the markup syntax directly, for that we suggest browsing the documentation at https://mystmd.org, instead we will focus our attention on the use cases for scientific publishing that we are trying to make as easy as possible. Specifically, the ability to add persistent identifiers (PIDs) and links to other structured content; hover previews to show details on demand; integrating live computational content and interactive figures; and exporting to many different formats including those used by scholarly publishing.
2.1Utility of Links and Identifiers¶
In MyST Markdown, citations can be added inline using @cite-key, following Pandoc-style syntax and referencing a BibTeX file in a project.
It is also possible to directly link to DOIs using @10.5281/zenodo.6476040, which will create a hover reference Cockett, 2022 as well as a references section at the bottom of the page.
This enhanced-links concept can be extended to Wikipedia, RRIDs, RORs, GitHub issues or code, and other scientific databases to augment writing.
For example, the link <rrid:SCR_008394> becomes RRID: SCR_008394, with rich-metadata and citations created.
Wikipedia links come with previews, for example, <wiki:gravitational_waves> becomes gravitational waves.
GitHub links to pull-requests also give hover information, for example, the following link #87 shows a hover preview in the online-version.
The use of DOIs and other structured scientific metadata can be reused in multiple different formats such as JATS XML and CrossRef deposits to create a DOI. Our goal with these integrations is to make the use of persistent identifiers (PIDs) both easy and rewarding to the author as they are writing. In traditional typesetting, this metadata is only added at publication time requiring specialized vendors and/or proprietary technology.
2.2Hover Previews for Content and Metadata¶
In Head et al. (2021) the authors show a speed up in comprehension of an article by 26% when showing information in context (i.e. “details on demand” Shneiderman, 1996), rather than requiring researchers to scroll back and forth to find figures and equations. MyST supports these concepts natively for cross-referencing equations, figures, and tables using hover-previews Example 1. This enhances the reading experience of scientific documents and retrieval of information.
In MyST Markdown we have also extended this “details on demand” concept to abbreviations to make it trivial to disambiguate the meaning of acronyms. In an analysis of over 18 million articles in Barnett & Doubleday (2020), the authors found that the vast majority of abbreviations (79%) appeared fewer than 10 times and many abbreviations had conflicting meanings even in the same discipline. In MyST Markdown, there is a trivial way to document abbreviations in YAML frontmatter or the project configuration See Program 1, these are then applied to all instances of that abbreviation in the article or notebooks giving a hover-preview and accessible HTML (e.g. try hovering over these in the online version: JATS, XML, VoR).
abbreviations:
UA: ulnar arteryProgram 1:YAML metadata used in MyST frontmatter to give accessible hovers to all abbreviations with minimal effort for the author. In this case, the acronym UA has over 18 distinct meanings in medicine European Association of Science Editors, 2019 not to mention other disciplines.
2.3Integrating Computational Content¶
Beyond structured typography, integrated metadata and hover-previews, MyST Markdown understands computational content and has been integrated with Jupyter Kluyver et al., 2016. The goal of this is two fold: (1) allowing for updates to computational content, figures, tables, and calculations to directly update documents; and (2) to bring interactive figures and integrated computation directly into articles. In the composition of a scientific narrative, scientists often use individual notebooks to create various components of their research (e.g. the preparation of a single figure, table or calculation). The outputs of these notebooks (a figure, table, or calculation) can then be used in a main, narrative-driven document — a scientific article or presentation.
In some cases, it is possible to collapse all computational information into a single article, and visually hide the code to focus on the narrative or presentational flow; an approach experimented with by eLife Tsang & Maciocci, 2020. This approach is appropriate for tutorials or the reproduction of visualizations rather than reproduction of a distinct detailed methodology that requires its own explanation and/or lengthy computation. Another approach is to include supplemental notebooks that can capture those individual steps e.g. Caprarelli et al., 2023, and transclude content Figure 2. Both approaches are appropriate in different circumstances, and depends on the goal of the communication, nature of the research, speed of execution, and if individual steps require dedicated narrative explanation. Additionally, the possibility of publishing a computational article, allows authors to rethink how to communicate their work and prepare specific visualizations and compact results datasets to take advantage of the format.
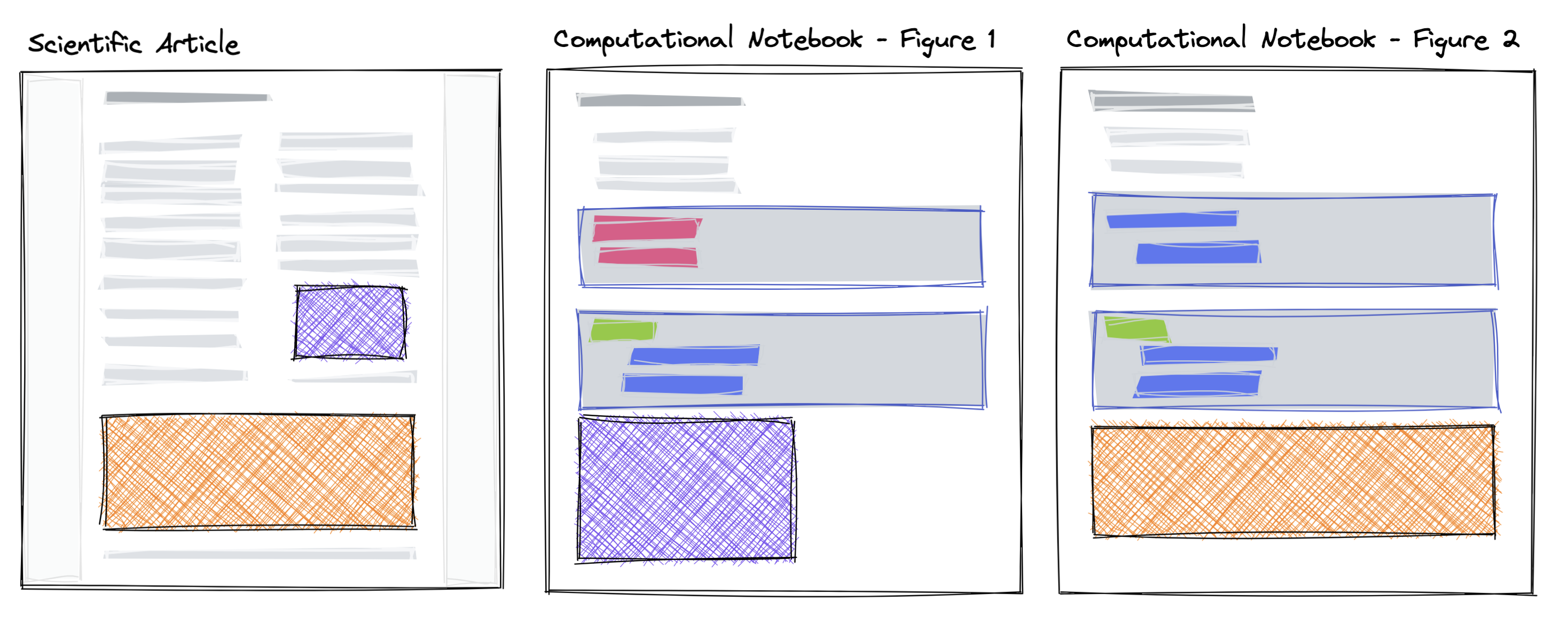
Figure 2:A schematic of embedding content from Jupyter Notebooks into an article. The purple and orange components, interactive figure or other computational outputs, are created in a computational notebook and subsequently used in other narrative or presentation-focused scientific article.
In Figure 2, we show an example of reusing computational outputs, such as figures or tables directly in a single computational research article. By embedding these rather than using a screenshot or copy-paste, any changes to the computational content can be immediately applied on a re-render. The embedding is completed through a simple MyST Markdown syntax that references a labeled cell in a Jupyter Notebook in, for example, a figure or table Program 2.
:::{figure} #embedded-cell
Additional caption.
:::Program 2:Embedding a cell from a supplementary notebook directly into a computational document by referencing the label/ID of the cell and adding a caption. The cell in the notebook must be labeled with a #| label: embedded-cell as the first line of the content.
Similar to the hover-references in Example 1, this approach improves the metadata around the notebooks and exposes individual outputs or code-snippets to be imported into other documents or projects. Additionally, we can attach either static-interactivity (e.g. Plotly, Altair) or dynamic computation (e.g. BinderHub, JupyterHub or JupyterLite) to these figures to run live computations directly in the article Example 2. Here we are aiming at a much richer, structured information commons that moves beyond just tracking scientific metadata towards easy-to-use tools that reuse scientific content.
2.4Single Source to Many Outputs¶
These capabilities of cross-references, typography and embedding visualizations and data-frames are complemented by a single-source export system that supports professional article templates and JATS XML Figure 4. This is referred to as single-source publishing e.g. Dunn, 2003, however, many implementations in scientific publishing focus first on manual translation to XML (e.g. from Word or LaTeX), rather than on an author-facing implementation.
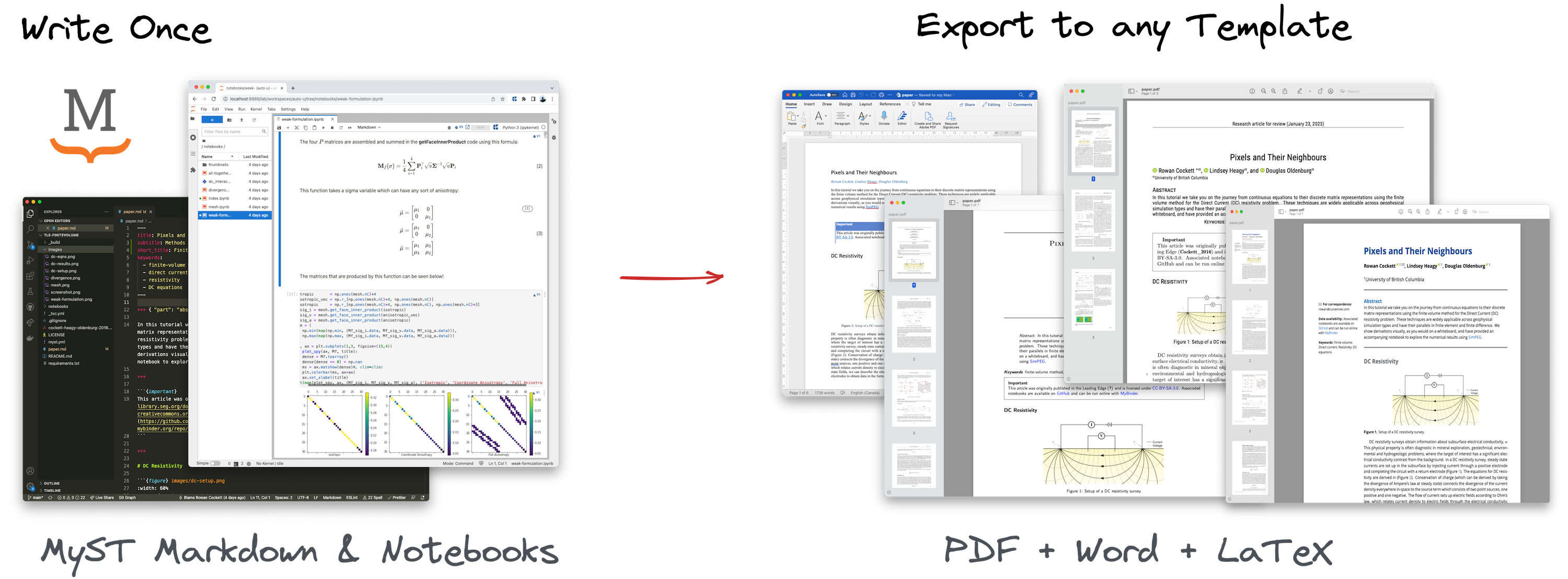
Figure 4:Export to PDF using LaTeX or Typst is supported for hundreds of different journal templates in addition to Microsoft Word or JATS XML, which is used throughout scientific publishing (showing content from Cockett et al. (2016); CC-BY-SA-4.0).
With single-source publishing, we can rely on rich transformations of the source content that can create professional PDFs, interactive HTML, and structured metadata such as JATS XML, which is the current standard for scientific archiving and text-mining.
3Publishing¶
The native way to publish or share a MyST article is through content-as-data, a collection of JSON files that represent the full documents and all associated metadata (if you are reading this online, you can add a .json to the URL to access the content).
This MyST content may be served alongside an independent website theme, which dynamically creates HTML based on these JSON files (very similar to XML-based single source publishing Dunn, 2003, but using modern web-tooling).
This server-side approach has a number of advantages, in that it allows you to maintain a journal/site theme, without having to upgrade or rebuild all content as would be required by static tooling.
It also puts content addressability as a first class concern (via the .json), enabling global cross referencing and opening up opportunities for varied publishing models in future.
This is the approach that we take with Curvenote journals, and provide managed services to maintain journal sites, manage and curate content, as well as provide the editorial management tools and workflows.
It is also possible to share MyST Markdown as a static HTML site using GitHub Pages.
To create a static site on GitHub pages you can run myst init --gh-pages, which will walk you through the steps of creating a GitHub action to publish your content statically.
In this scenario, a static HTML site is built from your content which can be hosted as any other static website, while some of the advantages of dynamic hosting are lost, it is an easy and accessible way for individuals to self-publish.
In 2024, Curvenote was asked to support the SciPy Proceedings and re-imagine a MyST based publishing approach that uses GitHub for open-peer-review, implementing a submission, editorial and peer review process with GitHub pull-requests. The original SciPy proceedings infrastructure was developed around 2010 and has influenced the design of the Journal of Open Source Software (JOSS), which has popularized GitHub-based peer-review Smith et al., 2018. In 2024, the workflow for SciPy Proceedings was updated to use MyST Markdown for the authoring process[12] (previously RST and LaTeX were supported, and the build process was in Sphinx), and the submission process now uses the open-source Curvenote CLI in combination with dedicated open-source Curvenote GitHub Actions to build, check, and preview the content of each commit, using GitHub workflows to automate the process, providing immediate feedback for authors and the conference editorial team.
3.1Structural Checks¶
The open-source Curvenote CLI (https://curvenote check in a MyST project will build the project and use the structured data to assess metadata (e.g. do authors provide ORCIDs), structural checks (e.g. does the article have an abstract?; is the article below a word count?), and check that references have valid DOIs.
We have designed these checks in a similar pattern to linting and/or unit tests in scientific software, which continually give feedback on the structural health of a codebase with near immediate feedback to authors Example 3.
Authors can take action to improve their metadata directly, by including DOIs, CRediT Roles, ORCIDs, and structural checks such as word count or missing sections.
For example, in the SciPy Proceedings in 2024, which used Curvenote checks for their submission system, required and optional metadata were improved by authors reacting to these automated checks without any intervention from the proceedings editorial team (e.g. scipy
3.2Automated Actions¶
Upon submission of there are a number of checks that are run and a submission or draft is deployed to Curvenote’s platform, which stores the structured MyST project.
We utilized GitHub Actions to automate initial checks and generate previews of submissions (via https://
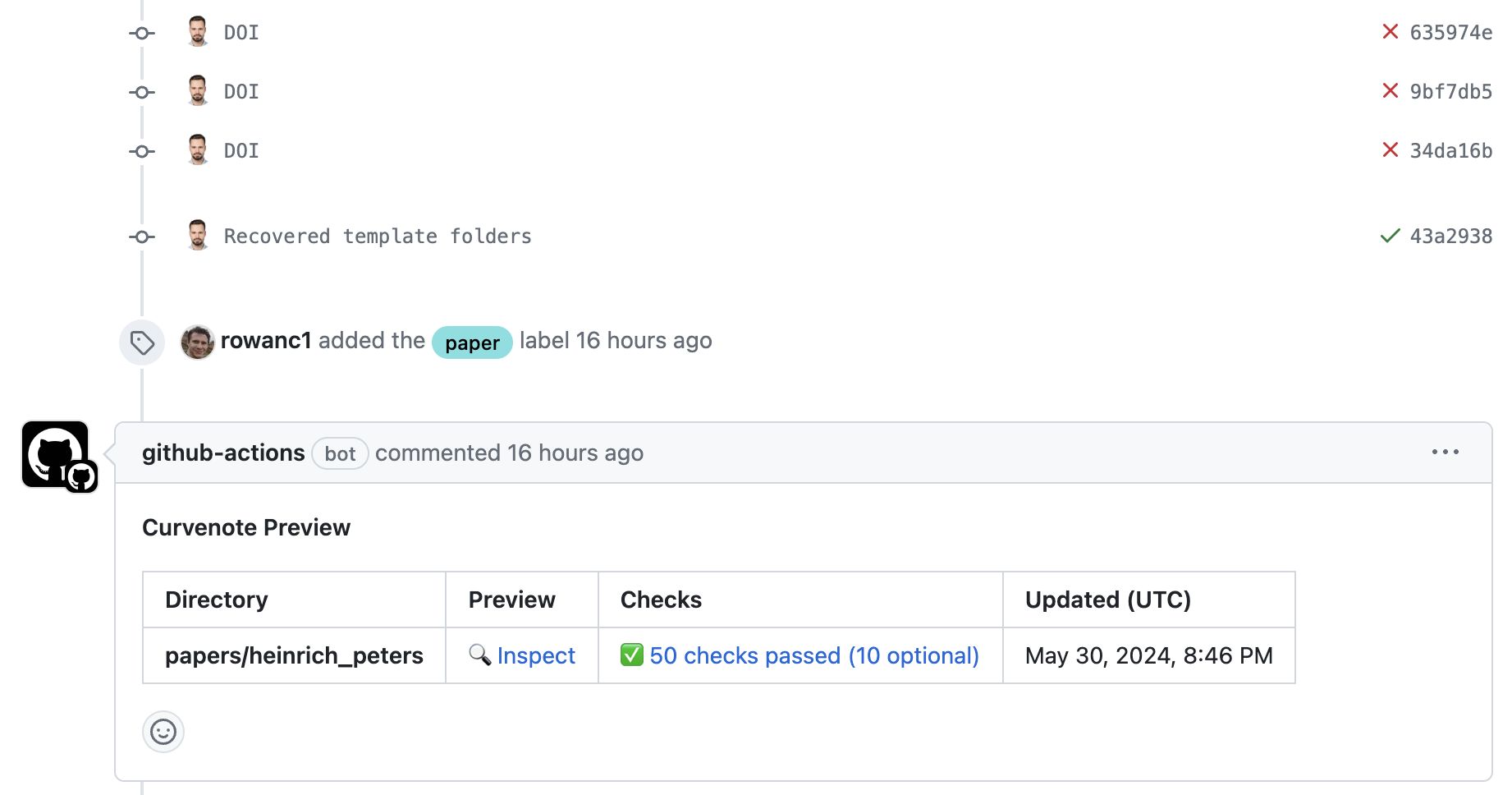
Figure 6:An example of a comment by a GitHub action, which shows the checks and preview of the article directly.
The checks in this example have promoted the author to improve metadata, see scipy
3.3Continuous Practices¶
MyST Markdown is a simple text-based format that can integrate directly with computational analysis and results in notebooks or scripts. This simplicity means a researcher’s manuscript can be easily created and maintained as an integral part of their research code base as they are working. This enables, for example, am article or draft that can be automatically rebuilt with the latest figures and data tables on every change, and any issues that you would hit on submission or publication are flagged as they are created. This is one of the ways that continuous science practices Section 1.3 can lead to radically improved efficiencies at scale.
Curvenote’s tools build on this theme of enabling researchers to have more visibility into how their work will appear when submitted and published.
The previews generated during the journal submission process, or by the actions on a researchers own repo can also be generated by an author at any time.
By running curvenote submit <journal> --draft researchers can get a set of check results and a preview in the style of that venue, and where submissions of Computational Articles are permitted, authors and reviewers will even get feedback on reproducible environment check that any interactive figures and notebooks execute as expected.
3.4Use Cases¶
The rapid feedback in authoring Section 2 coupled with structural checks Section 3.1, automation Section 3.2, and archiving of the content opens workflows for varied sizes of teams to adopt these continuous science practices. For example, individuals publishing static MyST Markdown sites, lab-groups creating a better archive to highlight the research contributions of their team (e.g. Applied Geophysics), conference proceedings (e.g. SciPy Proceedings), or more formalized society journals (e.g. Physiome, American Geophysical Union, Elemental Microscopy) Figure 7.

Figure 7:MyST Markdown and Curvenote tools can help lower the barrier to entry for scientific publishing workflows for journals, research institutes, conferences, private consortiums, universities, and lab groups.
4Conclusions¶
Scientific publishing infrastructure is disconnected from the day-to-day workflows of scientists, and at scale this slows the progress of science. These misalignments are particularly pronounced in computational disciplines, where rapid evolution of methodologies, software, and data demands equally dynamic and interconnected platforms. This gap — between the authoring/doing of research and the communicating/publishing of the research — slows the speed of research dissemination, reuse, and uptake and completely impedes “networked knowledge” and importing/reusing work in a structured way. For example, “importing” visualizations, equations or any other deeply-linked content – including provenance information – into new research articles, documentation or educational sites is completely impossible in today’s research ecosystem. As a metaphor, compare open-access science to open-source programming: it would be a world without package managers to share, version, reuse, and rapidly build upon other peoples work in a structured way. The open-source ecosystem would not exist without this infrastructure.
Open infrastructure for communicating science also has to be easy to integrate into existing tools, support computational, interactive components, be archivable for the long term, and be adopted by our existing sociotechnical system of societies, journals, and institutions. There are two interconnected problems that need to be solved: (1) upgrade existing scientific authoring tools, ensuring these are integrated into both scientific and data-science ecosystems; and (2) develop radically better ways to share content as individuals, small groups, preprints, and formalized, traditional journals with existing societies and institutions. The two problems are connected, in that the authoring tools should be able to deeply integrate with publishing mediums (e.g. referencing a figure from a publication should be able to show you that figure directly as you are authoring, including all interactivity and computation).
In this article we have presented some of the goals behind features of MyST Markdown and Curvenote, to support new open-science infrastructure including authoring tools as well as publishing workflows that support checks and automation. To support every next-generation research tool on top of open access knowledge, we need access to high-quality, structured content, data and software — not just the scholarly metadata and citation graph. Our goal of contributing to MyST Markdown is to make the processes behind creating this structured data more accessible and affordable. These tools in the hands of researchers can also enable process changes and continuous science practices: where checking and automation can support rapid iterations and feedback. The analogies between continuous delivery of software and continuous science give us an opportunity to peek ahead a decade to an analogous future and draw on many learnings on how to organize and focus infrastructure to get the best out of our scientific community.
There is, of course, an enormity of work ahead of these tools to transform science publishing at scale. We are grateful to our society partners who are changing community practices around publishing to support HTML-first publishing, experimenting with computational articles, and implementing new peer-review workflows. Tools on their own do not make change, but can help to enable it. Improvements to scientific publishing require many diverse community efforts to improve the quality and speed of how we communicate knowledge, and ultimately to accelerate scientific progress.
Portions of this manuscript have been previously shared in the Curvenote and MyST Markdown documentation.
The authors would like to thank Lindsey Heagy, Arfon Smith, Chris Holdgraf, Greg Caporaso, Jim Colliander, Fernando Perez, J.J. Allaire, and Kristen Ratan who have provided input on versions of these ideas over the last few years.
We would also like to thank the reviews Nate Jacobs, Angus Hollands, Lindsey Heagy, Stefan van der Walt, Andy Terrel, and Hongsup Shin who helped improved the manuscript.
Funding for portions of this work have come from the Sloan Foundation (for MyST Markdown and Notebooks Now!) and Alberta Innovates (for the initial version of the Curvenote CLI, which became mystmd).
Thank you to the growing list of MyST Markdown contributors who continue to make MyST a fantastic community project.
Copyright © 2024 Cockett et al. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license, which enables reusers to distribute, remix, adapt, and build upon the material in any medium or format, so long as attribution is given to the creators.
The Future of Research Communication and eScholarship (https://force11.org) conference, released their manifesto in 2012 Bourne et al. (2012) and much of that original writing still pertains to today, with the PDF being the main format of science communication.
A dispassionate observer, perhaps visiting from another planet, would surely be dumbfounded by how, in an age of multimedia, smartphones, 3D television and 24/7 social network connectivity, scholars and researchers continue to communicate their thoughts and research results primarily by means of the selective distribution of ink on paper, or at best via electronic facsimiles of the same.
MyST Markdown became a Jupyter Project on June 28, 2024 jupyter
/enhancement -proposals #123, and was previously hosted by Executable Books (https:// executablebooks .org). Curvenote is a company that provides many different tools for authoring and publishing content, including a collaborative WYSIWYG online editor that can export to MyST Markdown. In this article we discuss Curvenote’s open-source tools, specifically (a) a command-line interface (https://
github .com /curvenote /curvenote); and (b) GitHub actions for building and checking content (https:// github .com /curvenote /actions). We also highlight ideas from working with Curvenote’s partners when they pertain to improving scientific publishing. Direct publication costs include: checking of manuscript, copyediting, typesetting, formatting figures/graphs/tables, XML and metadata preparation, and handling corrections Grossmann & Brembs, 2021.
Global revenue in scientific publishing is around USD$19 billion, with over 50% of the market controlled by Elsevier, Wiley, Taylor & Francis, Springer Nature and SAGE Hagve, 2020.
The cost of transforming author submissions to produce structured content and metadata should approach zero, at least for a subset of users. For example, technical users who can use open-source command-line tools like MyST Markdown and GitHub. This has been shown to be the case for the Journal of Open Source Software (JOSS), for example, which advertizes a very low direct-publication cost of $2.71 per article Smith et al., 2018.
There are two types of feedback that we mean: (1) technical feedback as you are authoring, for example, “is this formatted correctly?” or “is this DOI correct?”; and (2) more substantial feedback from reviewers and readers who can only give you feedback when you have published. In the current system, technical feedback of an article-proof can take weeks and should be measured in milliseconds. Improving the immediacy of feedback from readers and peer-reviewers is a harder problem that involves how our existing sociotechnical system incentivizes article publishing rather than research communication and sharing findings as early and as often as possible.
In addition to increasing speed and robustness, continuous delivery practices also demonstrated that the high-performing teams spent 20% more time on new work, had 5-20% less manual work, and were 1.8x more likely to recommend their team as a great place to work DORA, 2018. These numbers are intriguing when contrasted to researchers, where (a) scientists already work 53.96 hours a week on average, and only about 36% of their time is actually spent on research (8% on grants, 32% on teaching, and 24% on service) Link et al., 2008; and (b) graduate students are six times more likely to experience depression than the general population Evans et al., 2018.
As an example of the the rapid and timely sharing of in-progress results and data, it is worth reflecting on the COVID-19 pandemic. Improved and more rapid sharing practices through data and preprints helped to develop a vaccine in record time Watson, 2022. Researchers published and shared data on the DNA sequence which enabled the design of a vaccine candidate in two days and the first manufacturing within two weeks.
By the second week of January 2020, researchers in China published the DNA sequence of SARS-CoV-2, the coronavirus that causes COVID-19. By early February, a COVID-19 vaccine candidate had been designed and manufactured.
In Massachusetts, the Moderna vaccine design took all of one weekend, and had been designed by January 13, two days after the genetic sequence had been made public.
In Penfold & Polka (2020), they describe the importance of adopting preprints, as the “overall peer review process can take years”. For the programmers reading this, it is worth a mental comparison to the pain of a pull-request being open for years. Having relevant feedback that is close to the time of implementation or writing is invaluable.
LaTeX is also supported by parsing and rendering directly with the
mystmdCLI, this is completed through@unified-latex.
- APC
- Article Processing Charge
- API
- Application Programming Interface
- CLI
- Command Line Interface
- CLOCKSS
- Controlled LOCKSS (Lots Of Copies Keep Stuff Safe)
- CRediT
- Contributor Roles Taxonomy
- DOI
- Digital Object Identifier
- HTML
- Hypertext Markup Language
- ID
- Identifier
- JATS
- Journal Article Tag Suite
- JOSS
- Journal of Open Source Software
- JSON
- JavaScript Object Notation
- LOCKSS
- Lots Of Copies Keep Stuff Safe
- MyST
- Markedly Structured Text
- NISO
- National Information Standards Organization
- ORCID
- Open Researcher and Contributor ID
- Portable Document Format
- PID
- Persistent Identifier
- REES
- Reproducible Execution Environment Specification
- ROR
- Research Organization Registry
- RRID
- Research Resource Identifier
- RST
- reStructuredText
- URL
- Uniform Resource Locator
- VoR
- Version of Record
- XML
- Extensible Markup Language
- YAML
- Yet Another Markup Language
- Bourne, P. E., Clark, T. W., Dale, R., de Waard, A., Herman, I., Hovy, E. H., & Shotton, D. (2012). Improving The Future of Research Communications and e-Scholarship (Dagstuhl Perspectives Workshop 11331). 10.4230/DAGMAN.1.1.41
- Wilkinson, M. D., Dumontier, M., Aalbersberg, Ij. J., Appleton, G., Axton, M., Baak, A., Blomberg, N., Boiten, J.-W., da Silva Santos, L. B., Bourne, P. E., Bouwman, J., Brookes, A. J., Clark, T., Crosas, M., Dillo, I., Dumon, O., Edmunds, S., Evelo, C. T., Finkers, R., … Mons, B. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data, 3(1). 10.1038/sdata.2016.18
- Grossmann, A., & Brembs, B. (2021). Current market rates for scholarly publishing services. F1000Research, 10, 20. 10.12688/f1000research.27468.2
- Hagve, M. (2020). Pengene bak vitenskapelig publisering. Tidsskrift for Den Norske Legeforening. 10.4045/tidsskr.20.0118
- Clotworthy, A., Davies, M., Cadman, T. J., Bengtsson, J., Andersen, T. O., Kadawathagedara, M., Vinther, J. L., Nguyen, T.-L., & Varga, T. V. (2023). Saving time and money in biomedical publishing: the case for free-format submissions with minimal requirements. BMC Medicine, 21(1). 10.1186/s12916-023-02882-y
- Xie, Y., Allaire, J. J., & Grolemund, G. (2018). R Markdown: The Definitive Guide. Chapman. 10.1201/9781138359444
- Project Jupyter, Bussonnier, M., Forde, J., Freeman, J., Granger, B., Head, T., Holdgraf, C., Kelley, K., Nalvarte, G., Osheroff, A., Pacer, M., Panda, Y., Perez, F., Ragan-Kelley, B., & Willing, C. (2018). Binder 2.0 - Reproducible, interactive, sharable environments for science at scale. Proceedings of the 17th Python in Science Conference. 10.25080/majora-4af1f417-011
- Caprarelli, G., Sedora, B., Ricci, M., Stall, S., & Giampoala, M. (2023). Notebooks Now! The Future of Reproducible Research. Earth and Space Science, 10(12). 10.1029/2023ea003458
- Tsang, E., & Maciocci, G. (2020). Welcome to a new ERA of reproducible publishing. ELife. https://elifesciences.org/labs/dc5acbde/welcome-to-a-new-era-of-reproducible-publishing
- Heidt, A. (2024). A publishing platform that places code front and centre. Nature. 10.1038/d41586-024-02577-1
- Mishra, A., & Otaiwi, Z. (2020). DevOps and software quality: A systematic mapping. Computer Science Review, 38, 100308. 10.1016/j.cosrev.2020.100308
- Leite, L., Rocha, C., Kon, F., Milojicic, D., & Meirelles, P. (2019). A Survey of DevOps Concepts and Challenges. ACM Computing Surveys, 52(6), 1–35. 10.1145/3359981
- Chen, L. (2015). Continuous Delivery: Huge Benefits, but Challenges Too. IEEE Software, 32(2), 50–54. 10.1109/ms.2015.27
- DORA. (2018). Accelerate: State of DevOps 2018: Strategies for a New Economy. Google Cloud. https://services.google.com/fh/files/misc/state-of-devops-2018.pdf
- Blinde, R. (2022). DevOps Unravelled: A Study on the Effects of Practices and Technologies on Organisational Performance [Master’s Thesis]. Leiden Institute of Advanced Computer Science (LIACS), Leiden University.
