
ITK-Wasm
Abstract¶
In recent years, WebAssembly (Wasm) has emerged as a widely-supported technology that offers high performance, compact binary size, support for multiple languages, hardware independence, security, and universal platform support, enabling developers to bring near-native speeds and portability to applications for the web and beyond. ITK-Wasm brings WebAssembly’s capabilities to scientific computing by combining the Insight Toolkit (ITK) and WebAssembly to enable high-performance spatial analysis across programming languages and hardware architectures.
In the scientific Python ecosystem, ITK-Wasm packages work in a web browser via Pyodide but also in system-level environments through the WebAssembly System Interface (WASI). ITK-Wasm bridges WebAssembly with scientific Python through simple, fundamental Python and NumPy-based data structures and Pythonic function interfaces. These interfaces can be accelerated through graphics processing units (GPU) or neural processing unit (NPU) implementations when available.
Beyond Python, ITK-Wasm’s integration of the WebAssembly Component Model launches scientific computing into a new world of interoperability, enabling the creation of accessible and sustainable multi-language projects that are easily distributed anywhere.
1Introduction¶
1.1Motivation¶
In the quest for enhanced interoperability and sustainability in scientific computing, WebAssembly (Wasm) emerges as a transformative technology. Wasm offers a universal, efficient compilation target, enabling high-performance computing across varied programming languages and hardware architectures Rossberg, 2019Rossberg, 2022Haas et al., 2017. This innovation is pivotal for scientific research, where analytical interoperability, tool sustainability, and computational efficiency are paramount. Wasm’s journey began with asm.js and evolved through Wasm and the WebAssembly System Interface (WASI).
1.2Brief history of Wasm¶
Asm.js was introduced by Alon Zakai via the Emscripten toolchain as a subset of JavaScript designed for high performance Zakai, 2011. It allowed developers to write code in languages like C and C++, compile it to asm.js, and run it in the browser with near-native performance. Asm.js achieved this by using a statically-typed subset of JavaScript that enabled optimizations by the JavaScript engine.
The predecessor to ITK-Wasm, ITK.js, supported compilation of C/C++ code into asm.js to enable reproducible, sustainable scientific computing in a web browser. An illustrative ITK.js application are interactive figures to replicate results from an open science article on an imaging denoising technique called anisotropic non-linear diffusion Mirebeau et al., 2014. Results on a simple webpage, hosted for free on GitHub Pages, are dynamically generated by the reader’s web browser, using the same code and data presented in the article. Presets load input data and set analysis parameters to dynamically reproduce the article’s figures. Additionally, readers can modify parameters to observe their effects or run the algorithm on their own image data.
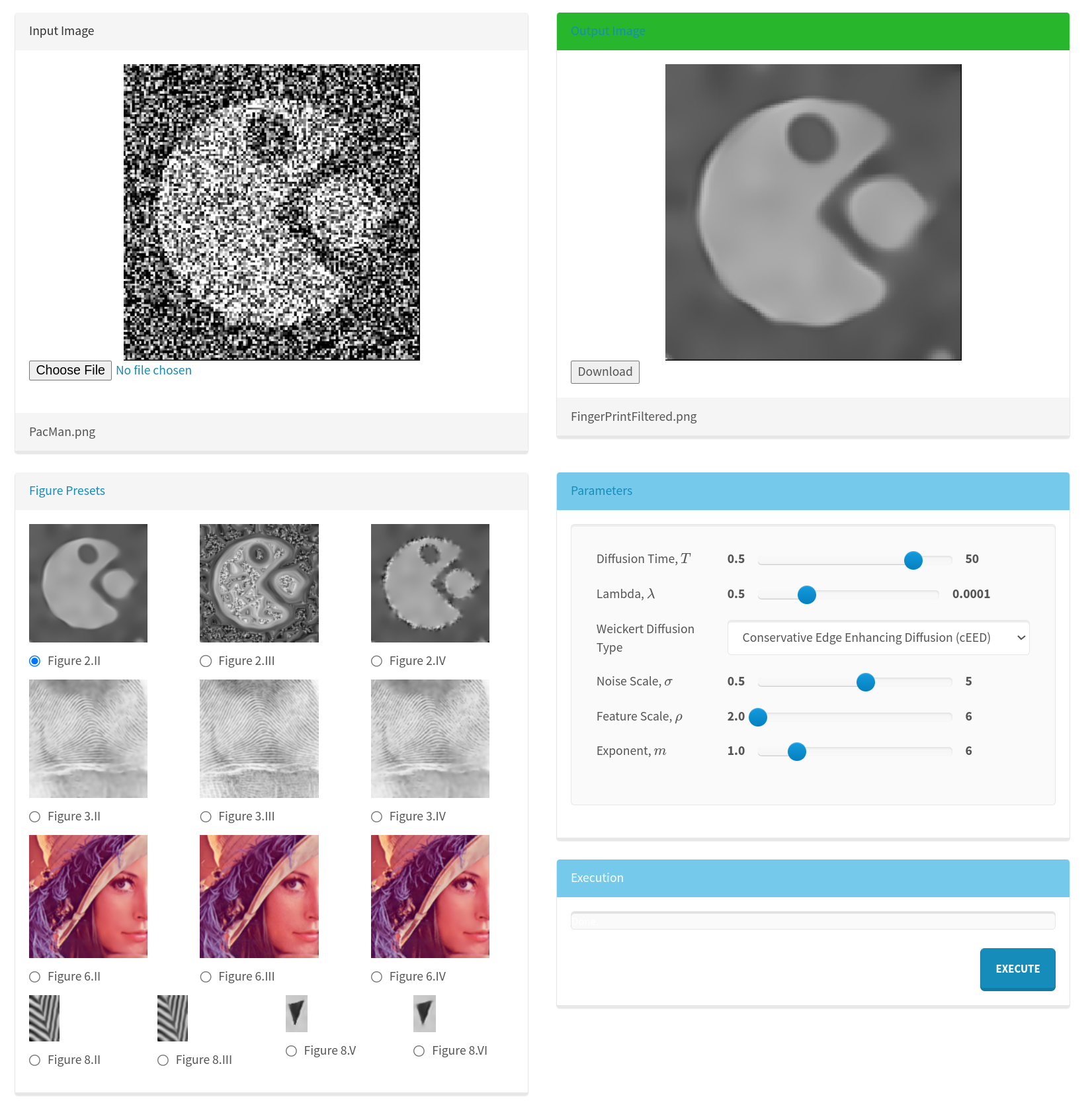
Figure 1:Open science interactive figures built with ITK.js, the predecessor to ITK-Wasm McCormick, 2015. The sustainability and accessibility of this interactive open science figure are remarkable, thanks to the utilization of open web technologies. These technologies, based on open standards, ensure compatibility across different systems and long-term support. This web application has continued to function flawlessly for over nine years without any maintenance, demonstrating its resilience and sustainability. It is hosted for free, with computations running directly on the reader’s system, eliminating the need for server-side resources. The application requires zero installation and can be accessed with just a few clicks, allowing users to easily reproduce results or apply the technique to their own data.
The key performance improvement with asm.js was its ability to utilize the JavaScript engine’s just-in-time (JIT) compilation to execute code faster than traditional JavaScript. However, asm.js had limitations, including verbose code and the overhead of JavaScript’s garbage collection and dynamic typing.
WebAssembly emerged from the limitations of asm.js. Announced in 2015 and reaching its initial MVP (Minimum Viable Product) in 2017, Wasm provides a compact binary format that could be executed at near-native speed Haas et al., 2017Rossberg et al., 2018. This innovation marked a significant leap in web performance, enabling complex applications to run efficiently in the browser.
Key features of Wasm include Rossberg et al., 2018:
Security
- Safe to execute
- Maintains the sandboxing paradigms of web browsers
Portability
- Language-, hardware-, and platform-independent
- Deterministic and easy to reason about
- Simple interoperability with the Web platform
Speed
- Fast to execute
- Maximally compact
- Easy to decode, validate and compile
- Easy to generate for producers
- Streamable and parallelizable
The WebAssembly System Interface (WASI), introduced by the WebAssembly Community Group, extends Wasm’s capabilities beyond the browser Dan Gohman et al., 2024. WASI provides a standardized system interface for WebAssembly, enabling it to interact with the underlying operating system. This development was crucial for running Wasm in desktop, server, and other non-browser environments.
Key features of WASI include:
- File system access
- Network access
- A modular architecture
With WASI, WebAssembly can be used to develop portable, high-performance applications that run anywhere. A plethora of WASI runtimes are available that vary in their focus, such as embedding in programming languages, specialized hardware such as field-programmable gate arrays (FPGAs), embedded devices, security, speed, or high performance computing (HPC) environments The WebAssembly System Interface (WASI), n.d.Chadha et al., 2023Zhang et al., 2024.
1.3Wasm and scientific computing¶
Throughout its evolution, WebAssembly has focused on performance improvements. Some notable advancements relevant to scientific computing include:
- Bulk memory operations: efficient copy and movement of data in memory
- SIMD support: Single Instruction, Multiple Data (SIMD) capabilities, allowing Wasm to perform parallel operations on multiple data points simultaneously with specialized instruction support available on modern CPUs
- Multithreading support: support for operating system threads and atomics for CPU parallelism
The evolution of WebAssembly from asm.js to Wasm to WASI has been marked by continuous improvements in performance, interoperability, and support for a wide range of programming languages and deployment environments. This journey has transformed WebAssembly into a versatile and powerful technology, capable of running high-performance applications anywhere, from the browser to the cloud.
Wasm has been embraced in commercial and industrial contexts for web applications, game development, edge computing, and server-side computing. However, its adoption in scientific computing has been more limited. This is partly due to the established reliance specialized software stacks in the scientific community. Additionally, the integration of Wasm into existing scientific workflows requires overcoming challenges related to data interoperability, toolchain compatibility, and the inertia of entrenched computational practices.
Enter ITK-Wasm, a pioneering resource that marries the Insight Toolkit (ITK) and open standards to seemlessly integrate Wasm for high-performance scientific spatial analysis or visualizationMcCormick, 2024McCormick et al., 2014Ibanez et al., 2024. ITK-Wasm supports both Emscripten-based Wasm in a web browser or WASI-SDK Wasm for system-level environments. ITK-Wasm is crafted to adhere to Wasm community standards, thereby facilitating the creation of Wasm modules that are simple, performant, portable, modular, and interoperable.
ITK-Wasm provides infrastructure that empowers research software engineers to:
- Build scientific C/C++ codes to Wasm
- Generate idiomatic programming language bindings, packages, and documentation
- Bridge Wasm with
- Local filesystems
- Canonical scientific programming data interfaces such as NumPy arrays
- Traditional scientific file formats with an emphasis on multi-dimensional spatial data
- Transfer data efficiently in and out of the Wasm runtime
- Support asynchronous and parallel execution of processing pipelines in way that is easy to understand and implement
2Methods¶
2.1Overview¶
ITK-Wasm provides powerful, joyful tooling for scientific computation in Wasm through a number of distinct but related parts.
- C++ core tooling
- Build environment Docker images
- A Node.js CLI to build Wasm, generate language bindings, run tests, and publish packages
- Small, language-specific libraries that facilicate idiomatic integration
- Scientific file format support
- Artificial intelligence and the semantic web integration
This tooling supports a straightforward programming model that aligns with functional programming paradigms and leverages Wasm’s simple stack-based virtual machine and Component Model architecture. All tooling is built around two key concepts:
- Interface Types: High-level, programming-language types for scientific computing, derived from Wasm’s low-level types.
- Processing Pipelines: Functions implemented in Wasm modules that operate on these interface types.
2.2C++ core¶
ITK-Wasm’s C++ core tooling provides:
- Fundamental numerical methods and multi-dimensional scientific data structures
- An elegant, modern interface to create processing pipelines
- A bridge to interoperable web techologies
- A bridge to Web3 and traditional desktop computing
These are embodied in the C++ core with:
- ITK
- CLI11
- Glaze
- libcbor
The Insight Toolkit (ITK) is an open-source, cross-platform library that provides developers with an extensive suite of software tools based on a proven, spatially-oriented architecture for processing scientific data in two, three, or more dimensions [McCormick et al. (2014);].
ITK includes fundamental numerical libraries, such as Eigen.
ITK’s C++ template-based architecture inherently helps keep Wasm modules small while enabling the compiler to add extensive performance optimizations.
The itk-wasm GitHub repository is also an ITK Remote Module, WebAssemblyInterface, that implements Wasm-interface specific functionality.
As an ITK Remote Module, wasm interface capabilities are available in an easily consumed library form.
To access wasm interface functionality in a build configuration,
find_package(ITK
COMPONENTS
WebAssemblyInterface
# Other desired components
)
include(${ITK_USE_FILE})
add_executable(a-wasm-pipeline ${srcs})
target_link_libraries(a-wasm-pipeline PUBLIC ${ITK_LIBRARIES})Glaze provides an elegent C++ JSON interface. This library was integrated as it is not only extremely fast but also small as a header-only library, which is critical for efficient WebAssembly deployment. The ability to read and write interface types to files, providing a bridge to Web3 and traditional desktop computing, is built on libcbor, which is another tiny footprint library.
Wasm module C++ processing pipelines are written with CLI11’s simple and intuitive argument parsing interface Henry Schreiner et al., 2024.
A C++ Wasm processing pipeline is defined with the familiar context of a command line executable.
Processing pipelines can be built with native binary toolchains into usable command line executables, which facilites development and debugging.
CLI11 extensions for the interface types and parsing enable an efficient embedding interface in addition to the command line interface.
All modules support an --interface-json flag that outputs a description of the module’s interface for binding generation.
2.3Build environment Docker images¶
Build environment Docker images encapsulate
- The ITK-Wasm C++ core
- An Emscripten or WASI toolchain
- Additional Wasm tools and configurations
These itkwasm/emscripten and itkwasm/wasi Docker images are dockcross images -- Docker images with pre-configured C++ cross-compiling toolchains that enable easy-application, reproducible builds, and a clean separation of the build environment, source tree, and build artifacts Developers, n.d..
These images include not only the CMake pre-configured toolchains, but pre-built versions of the ITK-Wasm C++ core. Moreover, Wasm tools for optimization, debugging, system execution, and testing are bundled. A number of build and system configurations are included to for optimized and debuggable builds.
2.4Command line interface (CLI)¶
An itk-wasm Node.js command line interface (CLI) drives
- Wasm module builds
- Generation of language bindings and language package configurations
- Testing for Wasm binaries
New projects are typically created with the create-itk-wasm CLI:
npx create-itk-wasmThe create-itk-wasm tool, which can be used interactively or programmatically, will generate C++ code with the required ITK-Wasm CLI11 interfaces, other support configuration files such as CMake build configuration scripts, a package.json file, language binding generation, and testing.
2.5Language-specific libraries and idiomatic bindings¶
Small, language-specific libraries are used by generated bindings to provide simple, clean, performant, and idiomatic interfaces in the host languages.
In TypeScript / JavaScript, this is the NPM itk-wasm package and in Python this is the PyPI itkwasm package.
TypeScript / JavaScript bindings and packages are generated from Emscripten toolchain builds of ITK-Wasm. TypeScript bindings are generated along with an NPM package.json to support package builds and deployment. Bindings are generated that support both browser-based execution and server-side execution in Node.js. Build scripts are provided to build TypeScript to JavaScript and also generate a demo app with an HTML interface. JavaScript bindings load Zstandard-compressed versions the Wasm modules on-demand in a web worker to support progressive and performant execution.
Python packages for new modules are generated for both system execution and web brower-execution. In the browser, Pyodide-compatible packages provide client-side web app scripting in Python, including via PyScript, and sustainable, scalable Jupyter deployments via JupyterLite. At a system level, Linux, macOS, and Windows operating systems are supported on x86_64 and ARM via wasmtime-py.
2.5.1Python environment dispatch¶
Bindings produce a primary, pip-installable Python package. In browser environments, this will pull a corresponding Emscripten-enabled Python package. For system Python distributions, this will bring in a corresponding WASI-enabled Python package. When GPU-accelerated implementations of functions are available in other packages along with required hardware and software, simply pip-installing the accelerator package will cause function calls to invoke accelerated overrides registered with modern package metadata, Figure 2.
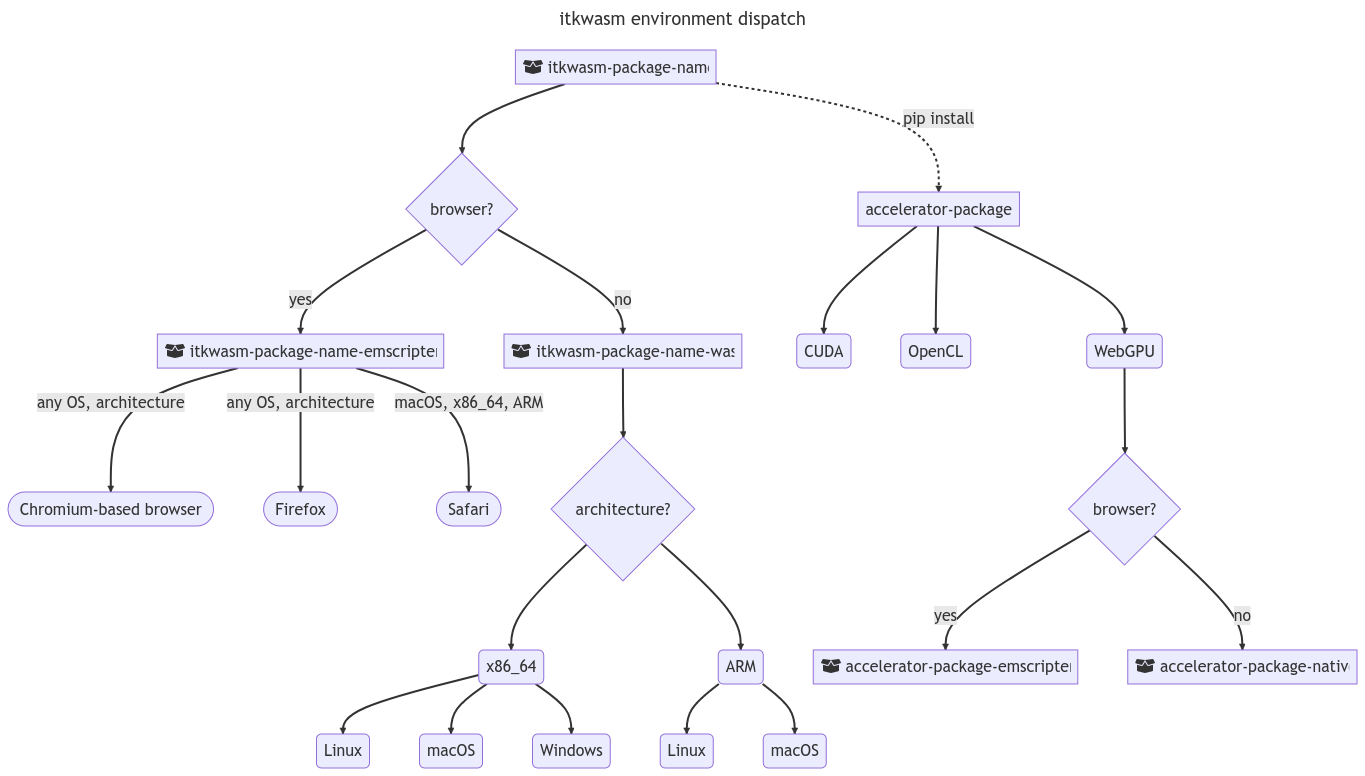
Figure 2:Cross-platform, cross-environment support with optional non-Wasm accelerator packages is made possible by a generated environment dispatch Python package, a WASI-based Python package, and a Pyodide package. The Pyodide package does not have typical Pyodide-Emscripten ABI limitations due to the application of principals from the Wasm Component Model.
2.5.2Browser and system APIs¶
While synchronous functions are available in system packages, browser packages provide asynchronous functions for non-blocking, performant execution in the JavaScript runtime event loop. These functions are called with modern Python’s async / await support.
For example, to install the itkwasm
pip install itkwasm-compress-stringifyIn Pyodide, e.g. the Pyodide REPL or JupyterLite,
import micropip
await micropip.install('itkwasm-compress-stringify')In the browser, call the async *_async function with the await keyword.
from itkwasm_compress_stringify import compress_stringify
data = bytes([33,44,55])
compressed = compress_stringify(data)from itkwasm_compress_stringify import compress_stringify_async
data = bytes([33,44,55])
compressed = await compress_stringify_async(data)2.6Traditional file format support¶
Assistance for handling data serialized in file formats plays a crucial role in enabling comprehensive analysis using a variety of software tools.
ITK-Wasm offers IO modules designed to interact with various standard scientific image and mesh file formats. These modules allow for the loading of data into the language-native interface type bindings.
Scientific image file formats supported include:
- AIM,ISQ
- BioRad
- BMP
- DICOM
- DICOM Series
- ITK HDF5
- JPEG
- GIPL (Guy’s Image Processing Lab)
- LSM
- MetaImage
- MGH
- MINC 2.0
- MRC
- NIfTI
- NRRD
- VTK legacy file format for images
- Varian FDF
Scientific mesh file formats supported include:
- BYU
- FreeSurfer surface, binary and ASCII
- OFF
- STL
- SWC Neuron Morphology
- OBJ
- VTK legacy file format for vtkPolyData
In addition to supporting external file formats, ITK-Wasm also introduces its own file formats. These ITK-Wasm file formats are optimized and offer a direct correspondence to spatial interface types, utilizing a straightforward JSON + binary array buffer format.
Execution pipeline WebAssembly modules only support ITK-Wasm formats by default -- this ensures that size of the WebAssembly pipeline binary is minimal. When using ITK-Wasm pipelines on with the command line interface, Wasm modules from the image-io and mesh-io packages can transform data in other formats into the format supported by all ITK-Wasm modules.
ITK-Wasm formats can be output in a directory or bundled in a single .cbor file. The Concise Binary Object Representation (CBOR) format supports JSON + binary array data in the data schema, is extremely small and lightweight in implementations, has support across programming languages, is highly performant, and provides a link to Web3 storage mechanisms.
ITK-Wasm file formats are available in ITK-Wasm IO functions but also in C++ via the WebAssemblyInterface ITK module. This module can be enabled in an ITK build by setting the -DModule_WebAssemblyInterface:BOOL=ON flag in CMake. And, loading and conversion is also available native-binary Python bindings via the itk-webassemblyinterface Python package.
2.7Artificial Intelligence and the Semantic Web¶
An ITK-Wasm LinkML Moxon et al., 2024 model provides FAIR definitions of the interface types that enable high-performance, portable, sustainable, and reproducible spatial analysis.
The interface types include:
- BinaryFile - Representation of a binary file on a filesystem. For performance reasons, use BinaryStream when possible, instead of BinaryFile.
- BinaryStream - Representation of a binary stream. For performance reasons, use BinaryStream when possible, instead of BinaryFile.
- Image - Representation of an N-dimensional scientific image.
- JsonCompatible - A type that can be represented in JSON.
- Mesh - Representation of an N-dimensional mesh.
- PointSet - Representation of a set of N-dimension points.
- PolyData - Representation of a polydata, 3D geometric data for rendering that represents a collection of points, lines, polygons, and/or triangle strips.
- TextFile - Representation of a text file on a filesystem. For performance reasons, use TextStream when possible, instead of TextFile.
- TextStream - Representation of a text stream. For performance reasons, use TextStream when possible, instead of TextFile.
- Transform - Representation of a parametric spatial transformation that can be applied to an Image, Mesh, PointSet, PolyData.
This model, combined with ITK-Wasm’s architecture, can perform analysis and visualization using natural language inputs provided to large-language artificial intelligence models. LinkML’s Pydantic models and TypeScript models enable interfaces like the Bioimage Chatbot Lei et al., 2023Lei et al., 2024 to semantically understand the needs of biologists without programming knowledge, allowing the system to execute desired operations or generate scripts for batch execution.
3Results¶
3.1Example application: generation of multiscale OME-Zarr images¶
Figure 3:Visible Male National Library of Medicine, 1994 frozen head computed tomography (CT) OME-Zarr volume, generated with ITK-Wasm. There are three resolution scales, which can be selected with the Image Scale buttons. Reduced resolutions are smoothed with a gaussian filter to avoid aliasing artifacts.
A notable application of ITK-Wasm is the generation of multiscale OME-Zarr images, a cloud-optimized bioimaging format with broad international adoption Moore et al., 2023Moore et al., 2021. To generate OME-Zarr’s multiscale representation of multidimensional bioimages, anti-aliasing filters must be applied.
Figure 4:A comparison of NRRD (Nearly Raw Raster Data), a traditional scientific image file format to the OME-Zarr scientific image file format, a modern, web-friendly file format. NRRD is a single, monolithic file with header and image pixel data formats concatenated. The header has a bespoke format and the pixel data is often compressed with the zlib compression codec. In contrast, OME-Zarr has a hierarchical structure, sometimes encoded in folders and files. Metadata is stored in JSON files. Image pixel data is stored in separate N-dimensional chunks that are compressed with fast, high compression ratio modern codecs like LZ4 or Zstd. Additionally, the image is represented at multiple resolutions. OME-Zarr’s chunk, highly-compressed, multiscale representation makes it ideal for uses cases like cloud-computing or extremely large images. However, this requires generation of the reduced resolution scales.
As detailed by the Nyquist-Shannon Sampling Theorem, high frequency content in an image must be reduced before downsampling to avoid aliasing artifacts.
Figure 5:Aliasing artifacts at the second resolution scale. With naive subsampling (top) aliasing artifacts introduce noise in the image at frequencies not supported by the sampling frequency. With gaussian anti-aliasing filtering prior to downsampling (bottom), signal fidelity is preserved.
In the NGFF-Zarr package Matt McCormick et al., 2024, ITK-Wasm anti-aliasing filters efficiently produce OME-Zarr images suitable for Pyodide, JupyterLite, traditional CPython environments, or analysis and visualization with other programming languages. While ITK-Wasm supports making general scientific C++ codes accessibly in wasm, in this example we will examine how the itk::DiscreteGaussianImageFilter is applied to address this problem Johnson et al., 2015Johnson et al., 2015McCormick et al., 2014.
We apply the N-dimensional gaussian convolution filter:
Where:
- is the Gaussian filter function
- is an n-dimensional vector representing spatial coordinates
- σ is the standard deviation of the Gaussian distribution
- is the number of dimensions
- is the squared Euclidean norm of
For downsampling, we use where is the downsampling factor, which is optimal Cardoso et al., 2015.
3.2C++ pipeline definition¶
The C++ wasm pipeline function, a pure function, is defined as a CLI11 executable Henry Schreiner et al., 2024. It uses an itk::wasm::Pipeline interface definition that operates on itk::wasm interface types.
int main(int argc, char * argv[])
{
itk::wasm::Pipeline pipeline("downsample", "Apply a smoothing anti-alias filter and subsample the input image.", argc, argv);
return itk::wasm::SupportInputImageTypes<PipelineFunctor,
uint8_t,
int8_t,
uint16_t,
int16_t,
uint32_t,
int32_t,
uint64_t,
int64_t,
float,
double
>
::Dimensions<2U, 3U, 4U, 5U>("input", pipeline);
}Here downsample defines the name of the pipeline function. A description for the pipeline is also provided -- this propagates to command line and language interface documentation.
In this function, we use the itk::wasm::SupportInputImageTypes utility to dispatch compile-time optimized pipeline based on the pixel type and dimension of the input image.
This ensures excellent performance while limiting the wasm module binary size, critical for performance and distribution, to the code that is used by the pipeline.
Next, pipeline inputs, outputs, and parameters are defined:
template<typename TImage>
class PipelineFunctor
{
public:
int operator()(itk::wasm::Pipeline & pipeline)
{
using ImageType = TImage;
constexpr unsigned int ImageDimension = ImageType::ImageDimension;
using InputImageType = itk::wasm::InputImage<ImageType>;
InputImageType inputImage;
pipeline.add_option("input", inputImage, "Input image")
->required()->type_name("INPUT_IMAGE");
std::vector<unsigned int> shrinkFactors { 2, 2 };
pipeline.add_option("-s,--shrink-factors", shrinkFactors, "Shrink factors")
->required()->type_size(ImageDimension);
std::vector<unsigned int> cropRadius;
pipeline.add_option("-r,--crop-radius", cropRadius, "Optional crop radius in pixel units.")
->type_size(ImageDimension);
using OutputImageType = itk::wasm::OutputImage<ImageType>;
OutputImageType downsampledImage;
pipeline.add_option("downsampled", downsampledImage, "Output downsampled image")
->required()->type_name("OUTPUT_IMAGE");
ITK_WASM_PARSE(pipeline);The types used are integers, floating point numbers, std containers of the same, or itk::wasm interface types. Long flags define parameter names in their language bindings, and their descriptions are propagated to their documentation.
The pipeline interface syntax can be generated from a set of interactive prompts provided by the create-itk-wasm CLI tool.
Once ITK_WASM_PARSE(pipeline) is called, input argument parsing and error handling is performed and the input pipeline options are populated with their values. During CLI execution, this means reading input files. When used with language bindings, files are not used and inputs are populated with in-memory content that was lowered into the wasm module with internal itk_wasm* functions.
Next comes the computational logic of the pipeline:
using GaussianFilterType = itk::DiscreteGaussianImageFilter<ImageType, ImageType>;
auto gaussianFilter = GaussianFilterType::New();
gaussianFilter->SetInput(inputImage.Get());
[...]
ITK_WASM_CATCH_EXCEPTION(pipeline, shrinkFilter->UpdateLargestPossibleRegion());
typename ImageType::ConstPointer result = shrinkFilter->GetOutput();
downsampledImage.Set(result);
return EXIT_SUCCESS;In this example, we are using ITK library C++ functionality, but this can be arbitrary C++ code.
The .Get() and .Set() methods on the interface types supply the C++ interface to the input values for computation and outputs. When the output interface type’s destructors are called, they are written to files on disk in a CLI context or prepared for wasm module lifting in an embedded language context.
The build is configured with simple, standard CMake:
cmake_minimum_required(VERSION 3.16)
project(itkwasm-downsample LANGUAGES CXX)
find_package(ITK REQUIRED
COMPONENTS
WebAssemblyInterface
ITKSmoothing
[...]
add_executable(downsample downsample.cxx)
target_link_libraries(downsample PUBLIC ${ITK_LIBRARIES})The same C++ and CMake code can be used for a native toolchain along with the wasm toolchain builds. This facilitates rapid development and easy debugging with native development tooling.
Additionally, CTest tests can be defined for native or WASI execution, e.g.:
enable_testing()
add_test(NAME downsample
COMMAND downsample
${CMAKE_CURRENT_SOURCE_DIR}/test/data/input/cthead1.png
${CMAKE_CURRENT_BINARY_DIR}/cthead1_downsampled.png
--shrink-factors 2 2
)In the WASI case, ITK-Wasm enables execution via a wasm interpreter and by enabling interpreter access to local input and output file directories.
3.3Command line invocation¶
If our example downsample pipeline module is built into a native binary, help output is generated with:
❯ ./downsample --helpThe equivalent invocation with the wasmtime wasm runtime for the WASI wasm module built from the same sources is:
❯ wasmtime run ./downsample.wasi.wasm --help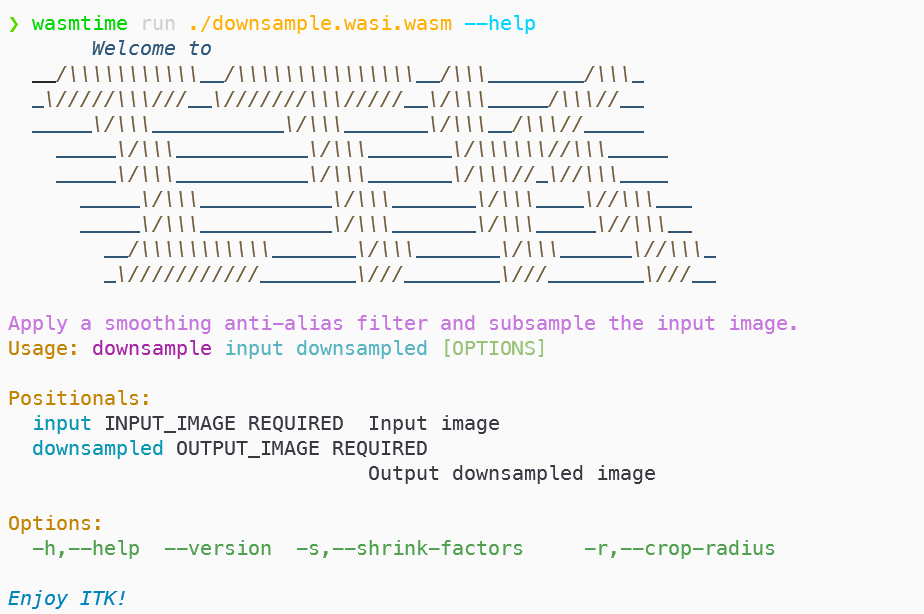
Figure 6:Wasm module help invocation and generated help output.
Where a native binary invocation is:
❯ ./downsample \
./vm_head.iwi.cbor ./downsampled.iwi.cbor \
--shrink-factors 4 4 2The equivalent wasm module invocation is:
❯ wasmtime run --dir=./ -- ./downsample.wasi.wasm \
./vm_head.iwi.cbor ./downsampled.iwi.cbor \
--shrink-factors 4 4 2All ITK-Wasm pipeline modules also support an --interface-json flag, which allows a module to self-describe its interface for documentation and language binding generation, described in the following sections.
❯ wasmtime run ./downsample.wasi.wasm --interface-json
{
"description": "Apply a smoothing anti-alias filter and subsample the input image.",
"name": "downsample",
"version": "0.1.0",
"inputs": [
{
"description": "Input image",
"name": "input",
"type": "INPUT_IMAGE",
"required": true,
"itemsExpected": 1,
"itemsExpectedMin": 1,
"itemsExpectedMax": 1,
"default": ""
}
],
"outputs": [
{
"description": "Output downsampled image",
"name": "downsampled",
"type": "OUTPUT_IMAGE",
"required": true,
"itemsExpected": 1,
"itemsExpectedMin": 1,
"itemsExpectedMax": 1,
"default": ""
}
],
"parameters": [
[...]
{
"description": "Shrink factors",
"name": "shrink-factors",
"type": "UINT",
"required": true,
"itemsExpected": 2,
"itemsExpectedMin": 2,
"itemsExpectedMax": 1073741824,
"default": "[2,2]"
},
[...]
]
}3.4Generating TypeScript packages from ITK-Wasm modules¶
One of the toolchains that ITK-Wasm supports is Emscripten. To facilitate seamless integration with modern web development, we generate TypeScript packages from Emscripten-generated wasm modules. These packages include Node.js bindings for server-side execution and browser-compatible interfaces for client-side execution.
3.4.1Node.js bindings¶
For server-side execution, our Node.js bindings provide direct access to the wasm module’s functionality. Local filesystem paths are made available to the wasm module, enabling pipelines to operate on files stored on the server. This allows developers to leverage ITK-Wasm pipelines in a Node.js environment, ideal for tasks such as image processing or analysis on a server.
3.4.2Browser bindings¶
In the browser, our bindings support pipelines that operate on files using the File object, as well as a custom BinaryFile object. The BinaryFile object consists of a string path and a Uint8Array binary, enabling efficient binary data transfer between the browser and wasm module. This allows developers to create web applications that interact with ITK-Wasm pipelines, enabling tasks such as image filtering and segmentation in the browser.
Figure 7:JavaScript and TypeScript package README rendered on npmjs.com.
3.4.3TypeScript Interface Types¶
Our TypeScript interface types are designed to be idiomatic classes comprised of JSON-compatible JavaScript types and TypedArrays. This ensures seamless interaction between the wasm module and TypeScript code, enabling developers to write efficient, natural, and type-safe code.
3.4.4WebWorker-based execution¶
To prevent interruption of the main user interface thread and support asynchronous background wasm compilation, pipelines are executed in a WebWorker when running in the browser. This ensures a responsive user experience while ITK-Wasm pipelines are executing.
3.4.5Parallelism with WebWorkerPool¶
For tasks that require parallel processing, a compatible WebWorkerPool is available. This enables developers to leverage multiple CPU cores to accelerate computationally intensive tasks, such as image registration and segmentation.
3.4.6Automated package configuration¶
From an ITK-Wasm project, we generate a complete TypeScript/JavaScript package configuration. This includes:
- TypeScript Bindings: Generated for both Node.js and browser environments.
- TypeScript Compilation Configuration: Pre-configured for optimal performance and compatibility.
- NPM Package Configuration: Ready for publication on the npm registry.
Figure 8:Interactive live API demo application.
3.4.7Documentation and demo app¶
To facilitate adoption and ease of use, we generate:
- README: A concise introduction to the project and its capabilities.
- Docsify Documentation: Detailed API documentation for the pipeline APIs.
- Demo App: An interactive testing environment for sample data or user-provided data, allowing developers to experiment with API parameters and visualize results.
By providing a comprehensive set of tools and configurations, we empower developers to harness the full potential of ITK-Wasm in modern web applications, streamlining the development of scientific imaging and analysis tools.
3.5Seamless Integration with Python Ecosystem¶
3.5.1Browser-based Pyodide Packages¶
We leverage the Emscripten toolchain to generate bindings for browser-based Pyodide Python packages. This enables seamless integration of ITK-Wasm with Pyodide, allowing developers to utilize ITK’s algorithms in web-based Python applications.
3.5.2Cross-Platform Compatibility with WASI¶
For system applications, we provide a WASI-based Python package that ensures cross-platform compatibility across all major platforms and architectures. This broadens the reach of ITK-Wasm, enabling developers to deploy ITK-based applications on a wide range of systems.
Figure 9:Downsample example package virtual environment size. ITK-Wasm WASI packages are simple, small, self-contained, and have minimal dependencies. The associated accessibility and sustainability for software that depends on these packages is reflected by the virtual environment size. When compared to a native binary or CUDA-based implementations, the WASI implementation has significantly reduced size and complexity.
3.5.3GPU Acceleration with cuCIM¶
To further enhance performance, we utilize the dispatch Python package’s capabilities in conjunction with a cuCIM accelerator package. This enables GPU acceleration, to enable improvements the execution speed, especially in applications where bulk data resides on the GPU, which is often the case for AI-enabled workflows.
Figure 10:Downsample example performance comparison between ITK-Wasm WASI, an equivalent ITK Python native binary implementation, and the ITK-Wasm CuCIM implementation. Executed on an Ryzen 9, 7940HS CPU, NVIDIA RTX 4070 Laptop GPU, Ubuntu 24.04 Linux system. Mean and standard deviation for ten iterations. While in this particular example the currently single-threaded WASI implementation is significantly slower than the native binary implementation, multi-threaded improvements with the native binaries hold promise for when this is enabled on the WASI binary (future work). NVIDIA CUDA-based ITK-Wasm CuCIM, applied without any other code changes when the itkwasm-downsample-cucim package is installed, demonstrates easy access to GPU acceleration when NVIDIA GPUs and CUDA software is available.
3.5.4API Documentation and Pythonic Interfaces¶
We generate API documentation for the simple, Pythonic interfaces, ensuring that developers can easily understand and utilize ITK-Wasm’s functionality. The interfaces are designed to be intuitive and easy to use, streamlining the development process.

Figure 11:Generated API documentation describes the Pythonic interfaces.
3.5.5Efficient Serialization for Parallel Computing¶
The interface types’ Python representation are built using Python data classes, comprising standard Python data types and NumPy arrays. This design enables trivial and efficient serialization, making it ideal for parallel computing with Dask. Developers can leverage this capability to scale their applications and tackle large-scale computing tasks.
3.5.6Broad Applicability in Scientific Computing¶
The utility of ITK-Wasm extends beyond web applications, as it can be seamlessly integrated into desktop applications like 3D Slicer Fedorov et al., 2012. This versatility demonstrates the broad applicability of ITK-Wasm in the scientific computing ecosystem, making it an invaluable tool for researchers and developers alike.
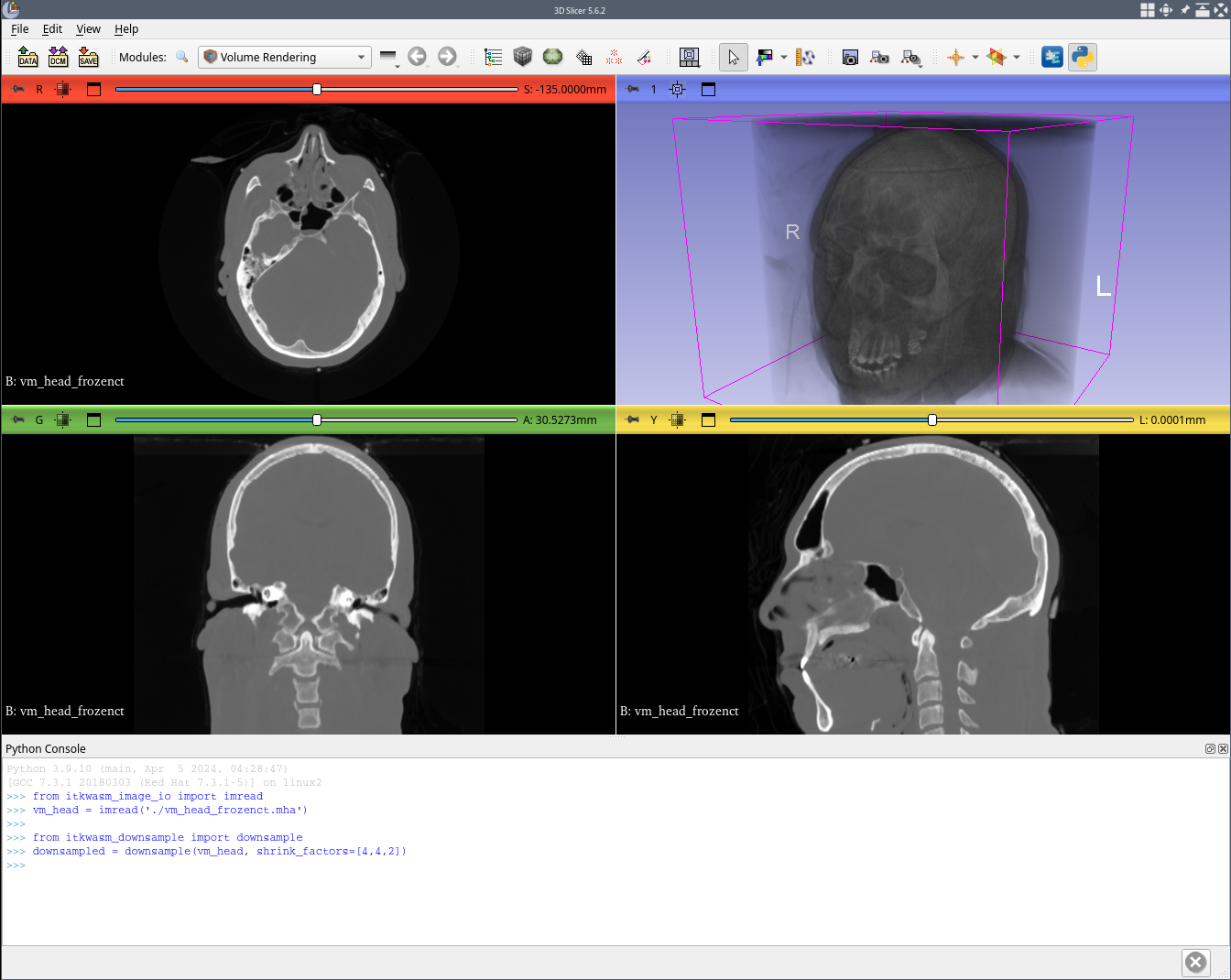
Figure 12:The itkwasm-downsample Python package in a traditional native desktop application, 3D Slicer.
4Discussion¶
WebAssembly was designed with interoperability in mind. Initially supporting languages like C and C++, the ecosystem has grown to include Rust, Go, Python, and more. This broad language support makes WebAssembly a versatile tool for developers across different domains.
ITK-Wasm’s approach, which focuses on bringing wasm’s capabilities to scientific software, excels in sustainability and composability thanks to small, self-contained, and idiomatic packages that are platform-agnostic and have minimal dependencies. This design enables outstanding computational reproducibility.
Future work will focus on enhanced integration of wasm community tools and standards:
- Multi-language Support: Support for bindings and package generation in additional languages like Java, C#, and Rust, broaden wasm module applicability.
- WebAssembly Interface Types: Standardizes the way Wasm modules interact with each other and with host environments, simplifying the integration process. We plan to bridge our interface types with the emerging Wasm Interface Type (WIT) definition.
- Component Model: An emerging standard that aims to improve modularity and reuse of Wasm components, further enhancing interoperability. Further instrumentation with the Component Model standard will enable generation of composite processing pipeline wasm modules that could be built from wasm component modules written in multiple languages.
ITK-Wasm provides a robust framework for scientific computing that leverages WebAssembly’s strengths. The framework bridges the gap between web-based and native applications, enabling high-performance, cross-platform scientific analysis. By integrating the principals of the WebAssembly Component Model, ITK-Wasm enhances interoperability and sustainability, allowing scientific Python to thrive in a multi-language ecosystem.
5Conclusion¶
ITK-Wasm stands at the forefront of fostering interoperability, multi-language program support, sustainability, accessibility, and reproducibility in scientific computing. By integrating the WebAssembly Component Model, ITK-Wasm not only enhances scientific Python’s capabilities but also sets a new standard for developing and distributing multi-language projects. The future of scientific computing is bright with ITK-Wasm’s contributions to the field, providing a universal platform for spatial analysis and visualization.
6Links:¶
- Documentation: https://
wasm .itk .org / - Source code: https://
github .com /InsightSoftwareConsortium /ITK -Wasm
The development of ITK-Wasm has been supported, in part, by the National Institute of Mental Health (NIMH) of the National Institutes of Health (NIH) under the BRAIN Initiative award number 1RF1MH126732.
Copyright © 2024 McCormick & Elliott. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license, which enables reusers to distribute, remix, adapt, and build upon the material in any medium or format, so long as attribution is given to the creators.
- API
- Application Programming Interface
- CLI
- Command Line Interface
- CPU
- Central Processing Unit
- FAIR
- Findable, Accessible, Interoperable, and Reusable
- FPGA
- Field-Programmable Gate Array
- GPU
- Graphics Processing Unit
- ITK
- Insight Toolkit
- JSON
- JavaScript Object Notation
- NPM
- Node Package Manager
- NPU
- Neural Processing Unit
- NRRD
- Nearly Raw Raster Data
- SIMD
- Single Instruction, Multiple Data
- WASI
- WebAssembly System Interface
- wasm
- WebAssembly
- Wasm
- WebAssembly
- Rossberg, A. (Ed.). (2019). WebAssembly Core Specification (1.0). W3C. https://www.w3.org/TR/wasm-core-1/
- Rossberg, A. (Ed.). (2022). WebAssembly Core Specification (2.0). W3C. https://www.w3.org/TR/wasm-core-2/
- Haas, A., Rossberg, A., Schuff, D. L., Titzer, B. L., Holman, M., Gohman, D., Wagner, L., Zakai, A., & Bastien, J. (2017). Bringing the web up to speed with WebAssembly. SIGPLAN Not., 52(6), 185–200. 10.1145/3140587.3062363
- Zakai, A. (2011). Emscripten: an LLVM-to-JavaScript compiler. Proceedings of the ACM International Conference Companion on Object Oriented Programming Systems Languages and Applications Companion, 301–312. 10.1145/2048147.2048224
- Mirebeau, J., Fehrenbach, J., Risser, L., & Tobji, S. (2014). Anisotropic Diffusion in ITK. The Insight Journal. 10.54294/en3833
- McCormick, M. (2015). Anisotropic Diffusion LBR. https://insightsoftwareconsortium.github.io/ITKAnisotropicDiffusionLBR/
- Rossberg, A., Titzer, B. L., Haas, A., Schuff, D. L., Gohman, D., Wagner, L., Zakai, A., Bastien, J. F., & Holman, M. (2018). Bringing the web up to speed with WebAssembly. Commun. ACM, 61(12), 107–115. 10.1145/3282510
- Dan Gohman, Lin Clark, Alex Crichton, Andrew Brown, Sam Clegg, Pat Hickey, Yosh, Dave Bakker, Colin Ihrig, YAMAMOTO Yuji, Peter Huene, Jakub Konka, Piotr Sikora, Chris Dickinson, Mike Frysinger, Robin Brown, YAMAMOTO Takashi, Syrus Akbary, Sergey Rubanov, … Nathaniel McCallum. (2024). WebAssembly/WASI: v0.2.2. Zenodo. 10.5281/ZENODO.4323446
- The WebAssembly System Interface (WASI). (n.d.). https://wasi.dev/
- Chadha, M., Krueger, N., John, J., Jindal, A., Gerndt, M., & Benedict, S. (2023). Exploring the Use of WebAssembly in HPC. Proceedings of the 28th ACM SIGPLAN Annual Symposium on Principles and Practice of Parallel Programming, 92–106. 10.1145/3572848.3577436
- Zhang, Y., Liu, M., Wang, H., Ma, Y., Huang, G., & Liu, X. (2024). Research on WebAssembly Runtimes: A Survey. http://arxiv.org/abs/2404.12621
- McCormick, M. (2024). itk-wasm: high-performance spatial analysis in a web browser, Node.js, and reproducible execution across programming languages and hardware architectures. Zenodo. 10.5281/ZENODO.3688880
- McCormick, M., Liu, X., Jomier, J., Marion, C., & Ibanez, L. (2014). ITK: enabling reproducible research and open science. Front. Neuroinform., 8, 13. 10.3389/fninf.2014.00013
- Ibanez, L., Lorensen, B., McCormick, M., King, B., Blezek, D., Johnson, H., Lowekamp, B., Jomier, J., Miller, J., Lehmann, G., Cates, J., Ng, L., Kim, J., Gelas, A., Zukić, D., Malaterre, M., Krishnan, K., Hoffman, B., Williams, K., … ITK Community Members. (2024). InsightSoftwareConsortium/ITK: ITK 5.4 Release Candidate 4: ALL THE DICOMs. Zenodo. 10.5281/ZENODO.889843
- Henry Schreiner, Philip Top, Chris Bachhuber, dherrera meta, Marcus Brinkmann, Andreas Deininger, Sam Hocevar, Rafi Wiener, Lucas Czech, Doug Johnston, Volker Christian, fpeng1985, Matt McCormick, polistern, captainurist, Sean Fisk, Ryan Curtin, Jose Luis Rivero, Jonas Nilsson, … Robert Adam. (2024). CLIUtils/CLI11: Version 2.4.2: Build systems. Zenodo. 10.5281/ZENODO.804964
