itk-elastix: Medical image registration in Python
Abstract¶
Image registration plays a vital role in understanding changes that occur in 2D and 3D scientific imaging datasets. Registration involves finding a spatial transformation that aligns one image to another by optimizing relevant image similarity metrics. In this paper, we introduce itk-elastix, a user-friendly Python wrapping of the mature elastix registration toolbox. The open-source tool supports rigid, affine, and B-spline deformable registration, making it versatile for various imaging datasets. By utilizing the modular design of itk-elastix, users can efficiently configure and compare different registration methods, and embed these in image analysis workflows.
Introduction¶
Image Registration¶
Image registration is a fundamental process in the field of scientific imaging that enables the alignment and comparison of images, facilitating the understanding of changes that occur within datasets. It involves finding a spatial transformation that optimizes relevant image similarity metrics, ensuring accurate alignment between images. A frequent registration type is the parametric approach where the spatial transformation is explicitly modeled. Examples of such transformation models are the rigid transform which allows translations and rotations, the affine transform that additionally includes shearing and the B-Spline transform that permits only local deformations. The reader can refer to Modersitzki Modersitzki, 2003 for an overview of the nonparametric registration. In addition to the parametric model, the choice of similarity metric plays a crucial role in the registration result and is dependent on the relationship of the pixel intensities between the images. Simple metrics such as normalized correlation are suitable for images with a linear intensity relationship, while more complex metrics such as mutual information Pluim et al., 2003 are employed for non-linear relationships.
Medical imaging heavily relies on image registration techniques Maintz & Viergever, 1998Oliveira & Tavares, 2014 to gain valuable insights and quantitative measurements. By registering medical images acquired at different time points or using various imaging modalities such as MRI and CT, researchers can analyze and quantify changes in anatomical structures, track disease progression and assess treatment efficacy. For instance, image registration allows the alignment of medical volumes across subjects to evaluate the impact of specific treatments, or the registration of sequential brain images to monitor tumor growth and response to therapy.
elastix¶
elastix Klein et al., 2009Shamonin et al., 2014 is a well-known and widely used open-source toolbox dedicated to image registration. It provides a comprehensive range of algorithms and utilities designed for aligning images using diverse transformation models, similarity measures, and optimization strategies. One of its key strengths is its modular design, enabling users to easily configure and combine different registration methods to suit application-specific needs. Parameter files govern the registration process by specifying transformation models, similarity measures, optimization strategies, and related parameters. By customizing these configurations, users can seamlessly adapt elastix to their specific requirements, ensuring optimal registration outcomes.
For example, when aligning an MRI brain scan with a CT scan using elastix, users can configure the transformation model, such as an affine or B-spline transformation model, to capture the geometric relationships between input images. They can also specify the similarity measure, like mutual information or normalized correlation, to evaluate the quality of alignment. Additionally, users have the flexibility to adjust optimization strategies, including parameters like the maximum number of iterations, to fine-tune the registration process and achieve optimal results. elastix supports both the more typical pairwise registration but also groupwise registration Metz et al., 2011Huizinga et al., 2016, where no image is specified as fixed but an implicit mean image is used instead as reference.
The elastix codebase is implemented in C++ and serves as an extension to the Insight Toolkit (ITK) McCormick et al., 2014. Through nearly two decades of development, elastix has achieved a mature state, characterized by stability, practical effectiveness, maintainability, and general backward compatibility. ITK Image data structures play a crucial role within elastix, representing multi-dimensional pixel data augmented with spatial information. Acting as a vital link between the digital pixel space and the physical space of the imaged object, ITK Images facilitate accurate registration. By computing transformations that map points from the physical space of one image to corresponding points in another, elastix achieves precise and meaningful alignment outcomes within the physical space. Complementing elastix, a utility software named transformix was developed to enable the application of registration results to additional images.
The original and still-supported method to utilize elastix and transformix are command line executables. For the end user, this approach has the advantage that it does not require any external dependencies to be installed, which eases deployment. However, one limitation of this executable-based approach is its reliance on file input/output (I/O) operations. To address this limitation and enable more efficient in-memory operations, a C++ API was developed for elastix and transformix. This API follows the paradigm established by ITK and its processing filters. By adopting this design approach, elastix and transformix gained the ability to perform operations directly in memory. This enhancement provides users with greater flexibility and efficiency in their image registration workflows.
To further accommodate the needs of the users in the continuously developing scientific computing ecosystem, wrappings of the C++ code to other languages was developed in the form SimpleElastix Marstal et al., 2016, which still exists as part of the SimpleITK Lowekamp et al., 2013 package. More recently, we have embarked on developing a Python-specific wrapper called itk-elastix. This wrapper extends the functionality of elastix and offers an ever-expanding collection of Jupyter Kluyver et al., 2016 examples, along with integration with other scientific processing libraries and visualization software. While there are other scientific python image image registration packages, itk-elastix stands out as a comprehensive Pythonic package with many image similarity metrics, implementations for 2D, 3D, and 4D images, and the ability to register a variety of imaging modalities. The subsequent sections of this paper delve into these aspects in greater detail.
itk-elastix: Python wrapping¶
The backend C++ elastix code is wrapped in Python with the Simplified Wrapper and Interface Generator (SWIG The SWIG development team, n.d.). The Python wrapping of elastix, itk-elastix, brings the power of elastix to the Python ecosystem, providing effortless integration with other scientific processing libraries and visualization software. The itk-elastix Python packages builds on the itk Python package’s pythonic interface and seamless integration with packages in the scientific Python ecosystem such as NumPy Harris et al., 2020. This enables users to leverage the rich functionality of elastix within their Python workflows, benefiting from its advanced image registration capabilities alongside popular Python libraries such as NumPy Harris et al., 2020, SciPy Virtanen et al., 2020, and MONAI Cardoso et al., 2022Diaz-Pinto et al., 2022.
The process of updating and distributing the itk-elastix Python package is as follows: Once a significant number of changes have been made to the C++ elastix repository, a pull request is initiated in the itk-elastix repository to update its version. This triggers the itk-elastix Continuous Integration (CI) system, which performs builds of Python packages across various Python versions (ranging from 3.7 to 3.11 at the moment of writing) and major platforms such as Windows, Linux, and macOS. When a git version tag is provided, the wrapped itk-elastix is automatically uploaded to PyPI, accompanied by a comprehensive summary of updates between the versions. As a result, users can easily install the latest itk-elastix by executing pip install itk-elastix within their Python environment. It is important to note that rigorous testing is conducted on the elastix backend functionality, with hundreds of tests performed during each pull request or commit, utilizing the CI system of the C++ repository. The test framework of elastix consists of various categories of tests, including low-level unit tests of the elastix library interface, minimal image registration tests on very small synthetic images, and larger regression tests of image registrations on realistic medical data. The tests are implemented using the CMake test driver CTest, the Python unittest module, and GoogleTest.
The Python wrapping for any ITK filter including elastix and transformix, offers two APIs: one functional and one object-oriented. We will describe the two API options and demonstrate the itk-elastix functionality with examples in the two following sections.
Functionality¶
Registration/transformation example¶
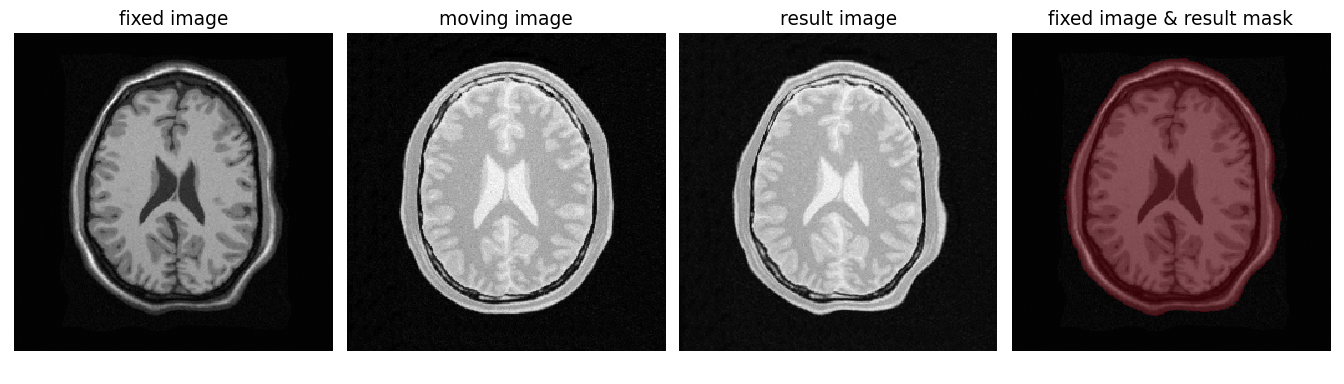
Figure 1:Synthetic example of 2D brain registration and transformation of masks.
The following example demonstrates the registration of 2D MRI brain images using the itk.elastix_registration_method and subsequent transformation of the corresponding moving mask using the itk.transformix_filter. The objective is to compare the overlap measure between the fixed mask and the transformed moving mask. It is important to note that this is a synthetic example where the fixed image intentionally exhibits significant deformations through an artificial non-linear transformation, solely for illustrative purposes. The masks utilized in this example represent segmentations of the head, including the brain and the skull. The procedure begins by reading the fixed and moving images from disk, followed by configuring a default set of B-spline registration parameters to be used for the registration process.
import itk
from scipy.spatial.distance import dice
# Load the moving and the fixed image from disk
fixed_image = itk.imread('./data/fixed.mha', itk.F)
moving_image = itk.imread('./data/moving.mha', itk.F)
# Configure a (default) parameter map with all the
# registration parameters
par_obj = itk.ParameterObject.New()
par_map = par_obj.GetDefaultParameterMap('bspline')
par_obj.AddParameterMap(par_map)
# Run the registration
# 1. The Object Oriented way
# elastix_obj = itk.ElastixRegistrationMethod.New(
# fixed_image,
# moving_image)
# elastix_obj.SetParameterObject(param_obj)
# elastix_obj.Update()
# result_image = elastix_obj.GetOutput()
# rtp = elastix_obj.GetTransformParameterObject()
# 2. The functional way
# rtp: result transform parameter object
result_image, rtp = itk.elastix_registration_method(
fixed_image,
moving_image,
parameter_object=par_obj)Following the registration process, we load the masks from disk and apply the transformation parameters obtained during registration to the moving mask. To preserve the binary nature of the masks and avoid introducing interpolation artifacts, we utilize the nearest neighbor interpolator. This choice ensures that the binary properties of the masks are maintained throughout the transformation process.
# Load the corresponding masks
fixed_mask = itk.imread('./data/f_mask.mha', itk.UC)
moving_mask = itk.imread('./data/m_mask.mha', itk.UC)
# Transform the moving mask using the result from the
# registration
rtp.SetParameter(0,
'ResampleInterpolator',
'FinalNearestNeighborInterpolator')
result_mask = itk.transformix_filter(moving_mask,
rtp)
# Compute dice on masks
initial_dice = 1 - dice(fixed_mask[:].ravel(),
moving_mask[:].ravel())
result_dice = 1 - dice(fixed_mask[:].ravel(),
result_mask[:].ravel())
print(initial_dice, result_dice)The last part of the code above calculates the Dice coefficient between the fixed mask and the transformed moving mask by converting the pixel arrays in the ITK Images into NumPy array views and then call scipy.distance.dice() on them. The initial Dice score was 97.88% which increased to 99.37% after registration. Figure 1 visualizes the fixed, moving and result image as well as an overlay of the fixed image and the transformed mask.
Jupyter Notebook collection¶
In addition to the core registration and transformation functionality demonstrated above, itk-elastix offers other additional features. To help new users who are starting out, and also keep existing users up-to-date with the new feature implementations, we offer an evolving collection of Jupyter Notebooks as usage examples. Each of the Notebooks covers usually a specific topic, can be run independently, and includes comments and detailed explanations. The Notebooks are also tested automatically by CI with each pull-request or commit, and hence it is ensured that they always reflect the current API and functionality of the codebase. Such Notebooks include, but are not limited to:
- specifying masks or point sets for the registration
- transforming point sets and meshes
- groupwise registration
- logging options
- saving output to disk options
- reading/writing transform in hd5f format
- calculation of spatial jacobian
- calculation of deformation field
- calculation of the inverse transform
- visualization of the registration
Interoperability with other packages¶
ITK Transforms¶
In addition to the fact that elastix is based on ITK, there is an ongoing effort to increase the compatibility between the two libraries even further. One particular example is the Transform classes Avants et al., 2012. In the following example, we show that ITK Transforms can be used directly by transformix:
# Create an ITK (translation) transform
transform = itk.TranslationTransform.New()
transform.SetOffset([50, -60])
# Specify the image space of the transform
sp = moving_image.shape
parameter_map = {
"Direction": ("1", "0", "0", "1"),
"Index": ("0", "0"),
"Origin": ("0", "0"),
"Size": (str(sp[1]), str(sp[0])),
"Spacing": ("1", "1")
}
par_obj = itk.ParameterObject.New()
par_obj.AddParameterMap(parameter_map)
# Pass an ITK transform directly to transformix
transformix_obj = itk.TransformixFilter.New(
moving_image)
transformix_obj.SetTransformParameterObject(par_obj)
transformix_obj.SetTransform(transform)
transformix_obj.Update()
# Get transformed (translated) image
translated_image = transformix_obj.GetOutput()NumPy & SciPy¶
Interoperability with NumPy and, consequently, with SciPy libraries, comes from functionality in ITK to convert ITK Images to NumPy arrays and vice versa. The relevant code is:
# itk image -> numpy array (deep copy)
image_array = itk.array_from_image(image_itk)
# itk image -> numpy array (shallow copy / view)
image_array = image_itk[:]
# numpy array -> itk image
image_itk = itk.image_from_array(image_array)Project MONAI¶
More and more people work on the application of deep learning to medical imaging research. To that end, we developed itk_torch_bridge as module of the MONAI codebase that allows conversion 1) of an ITK Image to a MONAI MetaTensor and the reverse, while making sure that all relevant metadata remain intact, and 2) an ITK Transform to a MONAI Transform and back. The latter is necessary since the ITK Transforms are defined in the world coordinate system while MONAI uses the pixel/voxel space. Example of a relevant application is performing deep learning registration (e.g. affine) using MONAI, and passing the Transform as initial Transform for itk-elastix, which can further register the images (e.g. non-linearly). Below, there is a short code snippet on how to use the module:
from monai.data import itk_torch_bridge as itb
import torch
# itk image <-> MONAI metatensor
image_mt = itb.itk_image_to_metatensor(image_itk)
image_itk = itb.metatensor_to_itk_image(image_mt)
# Transform: monai space <-> itk space
# affine_matrix: 3x3, matrix: 2x3, translation: 2x1
matrix, translation = itb.monai_to_itk_affine(
image=image,
affine_matrix=affine_matrix)Integration with other software¶
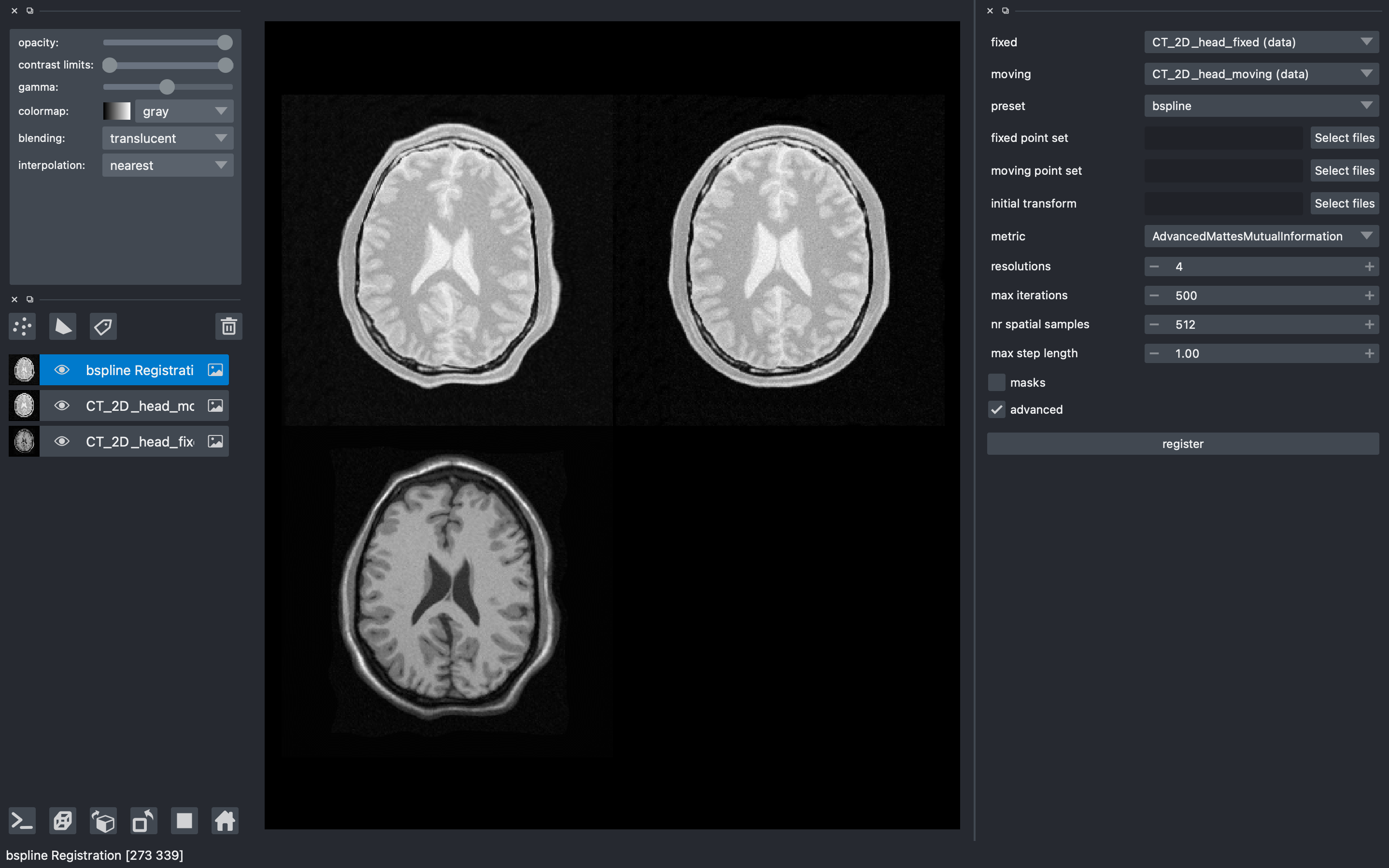
Figure 2:The user interface of the elastix-napari plugin. For a larger version of the image: https://
Continuous efforts have been made to make itk-elastix accessible to users of various tools. One notable community-driven initiative is SlicerElastix, which seamlessly integrates elastix (as an executable) into 3D Slicer Fedorov et al., 2012 medical image visualization software. In addition to this, recent endeavors focused on developing the elastix-napari plugin for the Napari napari contributors, 2019 visualization software, which is written in Python. Figure 2 illustrates Napari user interface and showcases an itk-elastix widget on the right side along with an example visualization of two input images and a transformed image at the center.
Documentation & reproducibility¶
elastix has been extensively used and cited for over a decade, resulting in the accumulation of significant community knowledge. In the spirit of reproducible science, and recognizing the value of building upon previous work, we have compiled a curated list of parameter files in a parameter file model zoo, each linked to its associated publication. This resource allows interested users to easily filter the list based on factors such as anatomical region, modality, or image dimensionality, empowering them to find pre-existing parameter files that suit their needs. By facilitating result replication on their own datasets and providing guidance for novel registration tasks, this initiative promotes reproducibility and collaboration within the community.
The documentation for each parameter, component, and API functionality is continuously updated using Sphinx, ensuring that it stays up-to-date with the latest developments in elastix. This allows users to access accurate and relevant information, with in-code descriptions automatically rendered as comments into a website for easy access and query capabilities. In addition, for a more comprehensive understanding of registration and the inner workings of elastix, the elastix manual provides in-depth descriptions covering various aspects, including detailed explanations of the algorithms and methodologies employed. To further support users, a community forum hosted as GitHub discussions serves as a valuable resource for asking questions, seeking assistance, and engaging in discussions with experienced users and developers who can provide support, share insights, and address any concerns or challenges faced by users.
Concluding remarks¶
We presented itk-elastix, an easy-to-install and easy-to-use Python package that lowers the entry barrier for multi-dimensional image registration. Its key features are 1) a robust and well-established backend codebase that provides stability and reliability, 2) an extensive collection of tutorials, a parameter file model zoo, and up-to-date documentation as comprehensive resources for user adoption, 3) seamless interoperability with popular scientific libraries in Python, including NumPy, SciPy, and MONAI, and 4) integration into 3D visualization software, facilitating visual analysis and interpretation of registered images. Overall, with itk-elastix, researchers and practitioners can effortlessly leverage the strengths of Python and seamlessly integrate it with a wide range of scientific software, which unlocks new possibilities and accelerates advancements in scientific image analysis. Next steps will further improve the applicability of itk-elastix on end-to-end deep learning segmentation and registration pipelines of diverse medical datasets. In addition, a port to WebAssembly will enhance the universal accessibility of the package.
Useful resources¶
- itk-elastix repository: https://
github .com /InsightSoftwareConsortium /ITKElastix - jupyter notebook examples: https://
github .com /InsightSoftwareConsortium /ITKElastix /tree /main /examples - elastix-napari plugin: https://
github .com /SuperElastix /elastix -napari - elastix community forum: https://
github .com /SuperElastix /elastix /discussions - parameter file model zoo: https://
elastix .lumc .nl /modelzoo / - elastix documentation and manual: https://
elastix .lumc .nl /doxygen /index .html
We gratefully acknowledge the financial support received from the Chan Zuckerberg Initiative (CZI) through the Essential Open Source Software for Science award for Open Source Image Registration: The elastix Toolbox, numbers 2020-218571 and 2021-237680 and the National Institute of Mental Health (NIMH) of the National Institutes of Health (NIH) under the BRAIN Initiative award number 1RF1MH126732.
Copyright © 2023 Ntatsis et al. This is an open-access article distributed under the terms of the Creative Commons Attribution 3.0 Unported license.
- API
- Application programming interface
- CI
- Continuous Integration
- CT
- computed tomography
- CZI
- Chan Zuckerberg Initiative
- ITK
- Insight Toolkit
- MRI
- magnetic resonance imaging
- NIMH
- National Institute of Mental Health
- OS
- operating system
- Modersitzki, J. (2003). Numerical methods for image registration. OUP Oxford.
- Pluim, J. P., Maintz, J. A., & Viergever, M. A. (2003). Mutual-information-based registration of medical images: a survey. IEEE Transactions on Medical Imaging, 22(8), 986–1004. 10.1109/JPROC.2003.817864
- Maintz, J. A., & Viergever, M. A. (1998). A survey of medical image registration. Medical Image Analysis, 2(1), 1–36. 10.1016/S1361-8415(01)80026-8
- Oliveira, F. P., & Tavares, J. M. R. (2014). Medical image registration: a review. Computer Methods in Biomechanics and Biomedical Engineering, 17(2), 73–93. 10.1080/10255842.2012.670855
- Klein, S., Staring, M., Murphy, K., Viergever, M. A., & Pluim, J. P. (2009). Elastix: a toolbox for intensity-based medical image registration. IEEE Transactions on Medical Imaging, 29(1), 196–205. 10.1109/TMI.2009.2035616
- Shamonin, D. P., Bron, E. E., Lelieveldt, B. P., Smits, M., Klein, S., Staring, M., & Initiative, A. D. N. (2014). Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Frontiers in Neuroinformatics, 7, 50. 10.3389/fninf.2013.00050
- Metz, C. T., Klein, S., Schaap, M., van Walsum, T., & Niessen, W. J. (2011). Nonrigid registration of dynamic medical imaging data using nD+ t B-splines and a groupwise optimization approach. Medical Image Analysis, 15(2), 238–249. 10.1016/j.media.2010.10.003
- Huizinga, W., Poot, D. H., Guyader, J.-M., Klaassen, R., Coolen, B. F., van Kranenburg, M., Van Geuns, R., Uitterdijk, A., Polfliet, M., Vandemeulebroucke, J., & others. (2016). PCA-based groupwise image registration for quantitative MRI. Medical Image Analysis, 29, 65–78. 10.1016/j.media.2015.12.004
- McCormick, M., Liu, X., Jomier, J., Marion, C., & Ibanez, L. (2014). ITK: enabling reproducible research and open science. Frontiers in Neuroinformatics, 8, 13. 10.3389/fninf.2014.00013
- Marstal, K., Berendsen, F., Staring, M., & Klein, S. (2016). SimpleElastix: A user-friendly, multi-lingual library for medical image registration. 2016 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), 574–582. 10.1109/CVPRW.2016.78
- Lowekamp, B. C., Chen, D. T., Ibáñez, L., & Blezek, D. (2013). The design of SimpleITK. Frontiers in Neuroinformatics, 7, 45. 10.3389/fninf.2013.00045
- Kluyver, T., Ragan-Kelley, B., Pérez, F., Granger, B., Bussonnier, M., Frederic, J., Kelley, K., Hamrick, J., Grout, J., Corlay, S., Ivanov, P., Avila, D., Abdalla, S., Willing, C., & development team, J. (2016). Jupyter Notebooks - a publishing format for reproducible computational workflows. In F. Loizides & B. Scmidt (Eds.), Positioning and Power in Academic Publishing: Players, Agents and Agendas (pp. 87–90). IOS Press. https://eprints.soton.ac.uk/403913/
- The SWIG development team. (n.d.). Simplified Wrapper and Interface Generator. https://www.swig.org/
- Harris, C. R., Millman, K. J., van der Walt, S. J., Gommers, R., Virtanen, P., Cournapeau, D., Wieser, E., Taylor, J., Berg, S., Smith, N. J., Kern, R., Picus, M., Hoyer, S., van Kerkwijk, M. H., Brett, M., Haldane, A., del Río, J. F., Wiebe, M., Peterson, P., … Oliphant, T. E. (2020). Array programming with NumPy. Nature, 585(7825), 357–362. 10.1038/s41586-020-2649-2
- Virtanen, P., Gommers, R., Oliphant, T. E., Haberland, M., Reddy, T., Cournapeau, D., Burovski, E., Peterson, P., Weckesser, W., Bright, J., van der Walt, S. J., Brett, M., Wilson, J., Millman, K. J., Mayorov, N., Nelson, A. R. J., Jones, E., Kern, R., Larson, E., … SciPy 1.0 Contributors. (2020). SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nature Methods, 17, 261–272. 10.1038/s41592-019-0686-2
