
libyt: a Tool for Parallel In Situ Analysis with yt
Abstract¶
In the era of exascale computing, storage and analysis of large scale data have become
more important and difficult.
We present libyt, an open source C++ library, that allows researchers to analyze and
visualize data using yt or other Python packages in parallel during simulation runtime.
We describe the code method for organizing adaptive mesh refinement grid data structure and
simulation data, handling data transition between Python and simulation with minimal memory
overhead, and conducting analysis with no additional time penalty using Python C API and
NumPy C API.
We demonstrate how it solves the problem in astrophysical simulations and increases disk
usage efficiency. Finally, we conclude it with discussions about libyt.
Introduction¶
In the era of exascale computing, storage and analysis of large-scale data has become a critical
bottleneck.
Simulations often use efficient programming language like C and C++, while many data analysis tools
are written in Python, for example yt[1] Turk et al., 2011.
yt is an open-source, permissively-licensed Python package for analyzing and visualizing
volumetric data.
It is a light weight tool for quantitative analysis for astrophysical data, and it has also been
applied to other scientific domains.
Normally, we would have to dump simulation data to hard disk first before conducting analysis using
existing Python tools.
This takes up lots of disk space when the simulation has high temporal and spatial resolution.
This also forces us to store full datasets, even though our region of interest might contain only
a small portion of simulation domain.
It makes large simulation hard to analyze and manage due to the limitation of disk space.
Is there a way to probe those ongoing simulation data using robust Python ecosystem?
So that we don’t have to re-invent data analysis tools and solve the disk usage issue at the same
time.
In situ analysis, which is to analyze simulation data on-site, without intermediate step of writing
data to hard disk is a promising solution.
It also reduces the barrier of analyzing data by using well-developed tools instead of creating our own.
We introduce in situ analysis tool libyt[2] , an open source C++ library, that allows researchers to analyze and
visualize data by directly calling yt or any other Python packages during simulations runtime
under parallel computation.
Through wrapping ongoing simulation data using NumPy C API Harris et al., 2020, constructing proper Python
C-extension methods and Python objects using Python C API Van Rossum & Drake, 2009, we can reuse C++ runtime
data and realize back-communication of simulation information, allowing user to define their own
data generating C function, and use it to conduct analysis inside Python ecosystem.
This is like using a normal Python prompt, but with direct access to simulation data.
libyt provides another way for us to interact with simulations.
In this proceeding, we will describe the methods in Section Code Method, demonstrate how libyt
solve the problem in Section Applications, and conclude it with Section Discussions.
Code Method¶
Overview of libyt¶
libyt serves as a bridge between simulation processes and Python instances as
illustrated in Figure 1.
It is the middle layer that handles data IO between simulations and Python instances,
and between MPI processes.
When launching N MPI processes, each process contains one piece of simulation and
one Python interpreter. Each Python interpreter has access to simulation data.
When doing in situ analysis, every simulation process pauses, and a total of N Python
instances will work together to conduct Python tasks in the process space of MPI.
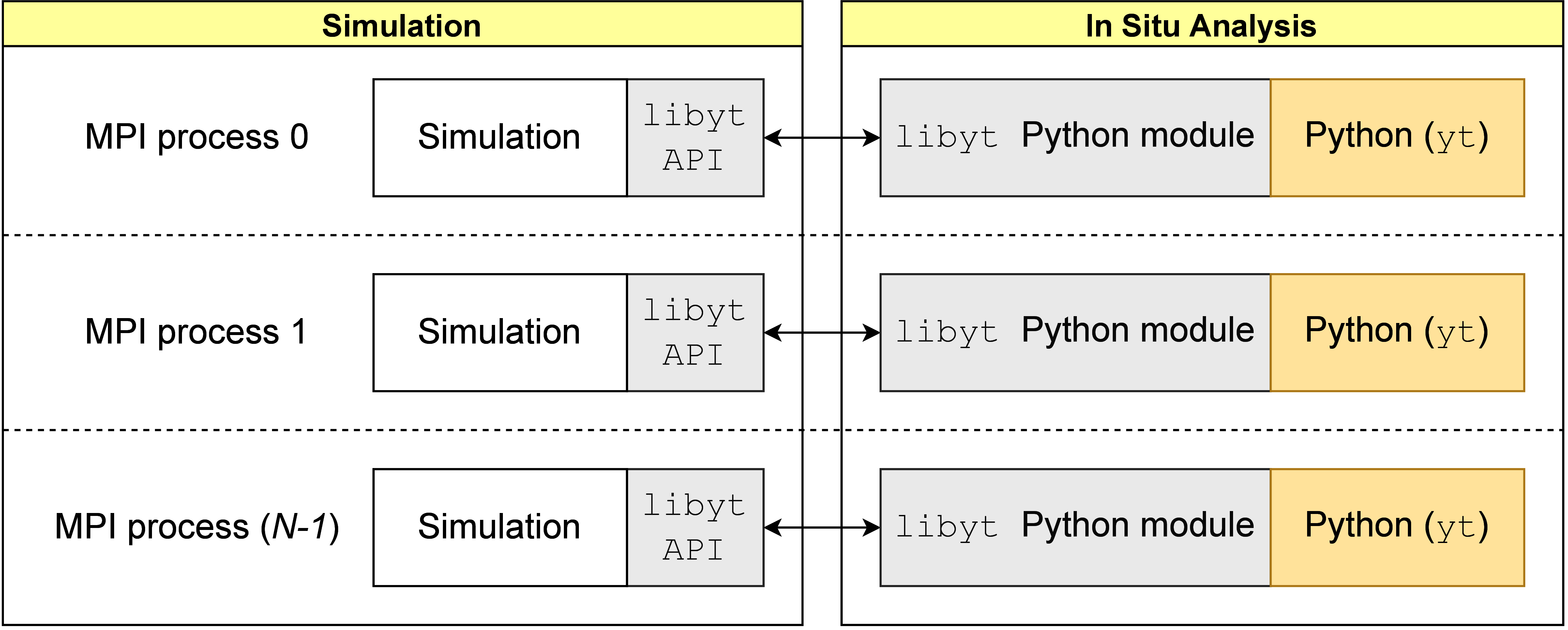
Figure 1:This is the overall structure of libyt, and its relationship with simulation
and Python. It provides an interface for exchanging data between simulations and
Python instances, and between each process, thereby enabling in situ parallel
analysis using multiple MPI processes. libyt can run arbitrary Python scripts
and Python modules, though here we focus on using yt as its core analysis
platform.
Simulations use libyt API[3] to pass in data and run Python codes during runtime,
and Python instances use libyt Python module to request data directly from simulations
using C-extension method and access Python objects that contain simulation information.
Using libyt for in situ analysis is very similar to running Python scripts in post-processing
under MPI platform, except that data are stored in memory instead of hard drives.
libyt is for general-purpose and can launch arbitrary Python scripts and Python modules,
though here, we focus on using yt as our core analysis tool.
Connecting Python and Simulation¶
We can extend the functionality of Python by calling C/C++ functions, and, likewise, we can also embed Python in a C/C++ application to enhance its capability. Python and NumPy provides C API for users to connect objects in a main C/C++ program to Python.
Currently, libyt supports only adaptive mesh refinement (AMR) grid data structure.[4]
How libyt organizes simulation with AMR grid data structure is illustrated in Figure 2.
It first gathers and combines local adaptive mesh refinement grid information
(e.g., levels, parent id, grid edges, etc) in each process, so that every Python instance contains
full information.
Next, it allocates array using PyArray_SimpleNew and stores the information in a linear
fashion according to global grid id.
The array can be easily looked up, and we can retrieve information by libyt at C side using PyArray_GETPTR2.
The operation only involves reading elements in an array.
The array is accessible both in C/C++ and Python runtimes.
For simulation data, libyt wraps those data pointers using NumPy C API PyArray_SimpleNewFromData.
This tells Python how to interpret block of memory (e.g., shape, type, stride) and does not make a copy.
libyt also marks the wrapped data as read-only[5] to avoid Python accidentally alters it,
since they are actual data used in simulation’s iterative process.
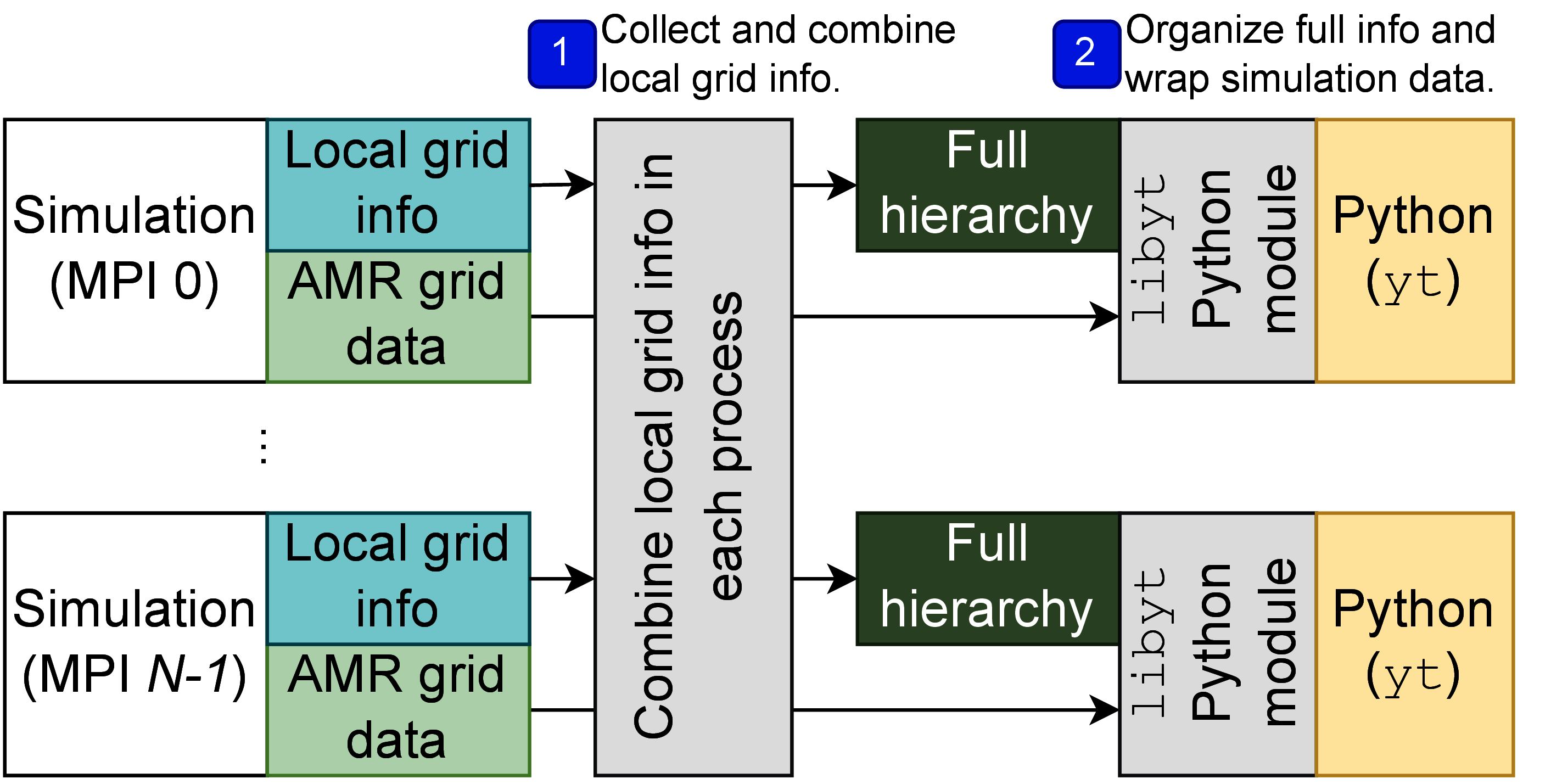
Figure 2:This diagram shows how libyt loads and organizes simulation information and
data that is based on adaptive mesh refinement (AMR) grid data structure.
libyt collects local AMR grid information and combines them all, so that each
Python instance contains whole information.
As for simulation data, libyt wraps them using NumPy C API, which tells Python
how to interpret block of memory without duplicating it.
libyt also supports back-communication of simulation information.
Figure 3 shows the mechanism behind it.
The process is triggered by Python when it needs the data generated by a user-defined
C function. This usually happens when the data is not part of the simulation iterative
process and requires simulation to generate it, or the data isn’t stored in a contiguous
memory block and requires simulation to help collect it.
When Python needs the data, it first calls C-extension method in libyt Python module.
The C-extension method allocates a new data buffer and passes it to user-defined C function,
and the function writes data in it.
Finally, libyt wraps the data buffer and returns it back to Python.
libyt makes the data buffer owned by Python[6], so that the data gets freed when it is no
longer needed.
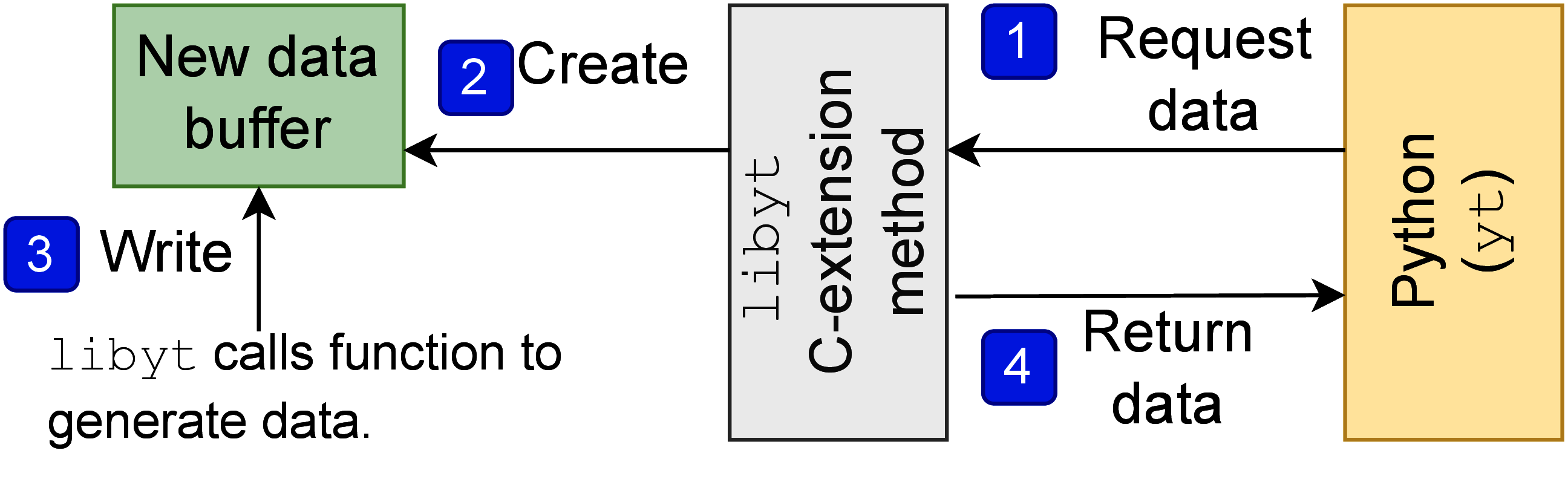
Figure 3:This diagram describes how libyt requests simulation to generate data using
user-defined C function, thus enabling back-communication of simulation information.
Those generated data is freed once it is no longer used by Python.
Grid information and simulation data are properly organized in dictionaries under libyt
Python module. One can import it during simulation runtime:
import libyt # Import libyt Python moduleIn Situ Analysis Under Parallel Computing¶
Each MPI process contains one simulation code and one Python instance.
Each Python instance only has direct access to the data on local computing nodes,
thus all Python instances must work together to make sure everything is in reach.
During in situ Python analysis, workloads may be decomposed and rebalanced according
to the algorithm in Python packages.
It is not necessary to align with how data is distributed in simulation.
Even though libyt can call arbitrary Python modules, we focus on how it uses yt and MPI
to do analysis under parallel computation here.
yt supports parallelism feature[7] using mpi4py[8] as communication method.
libyt borrows this feature and utilizes it directly.
The way yt calculates and distributes jobs to each MPI process is based on data locality,
but it does not always guarantee to do so[9].
In other words, in in situ analysis, the data requested by yt in each MPI process does not
always locate in the same process.
Furthermore, there is no way for libyt to know what kind of communication pattern a Python script needs in advance.
For a much more general case, it is difficult to schedule point-to-point communications that fit
any kind of algorithms and any number of MPI processes.
libyt uses one-sided communication in MPI, also known as Remote Memory Access (RMA),
by which one no longer needs to explicitly specify senders and receivers.
Figure 4 describes the data redistribution process in libyt.
libyt first collects requested data in each process and asks each process to prepare it.
Then libyt creates an epoch, for which all MPI processes will enter, and each process can fetch the data
located on different processes without explicitly waiting for the remote process to respond.
The caveat in data exchanging procedure in libyt is that it is a collective operation, and requires every
MPI process to participate.
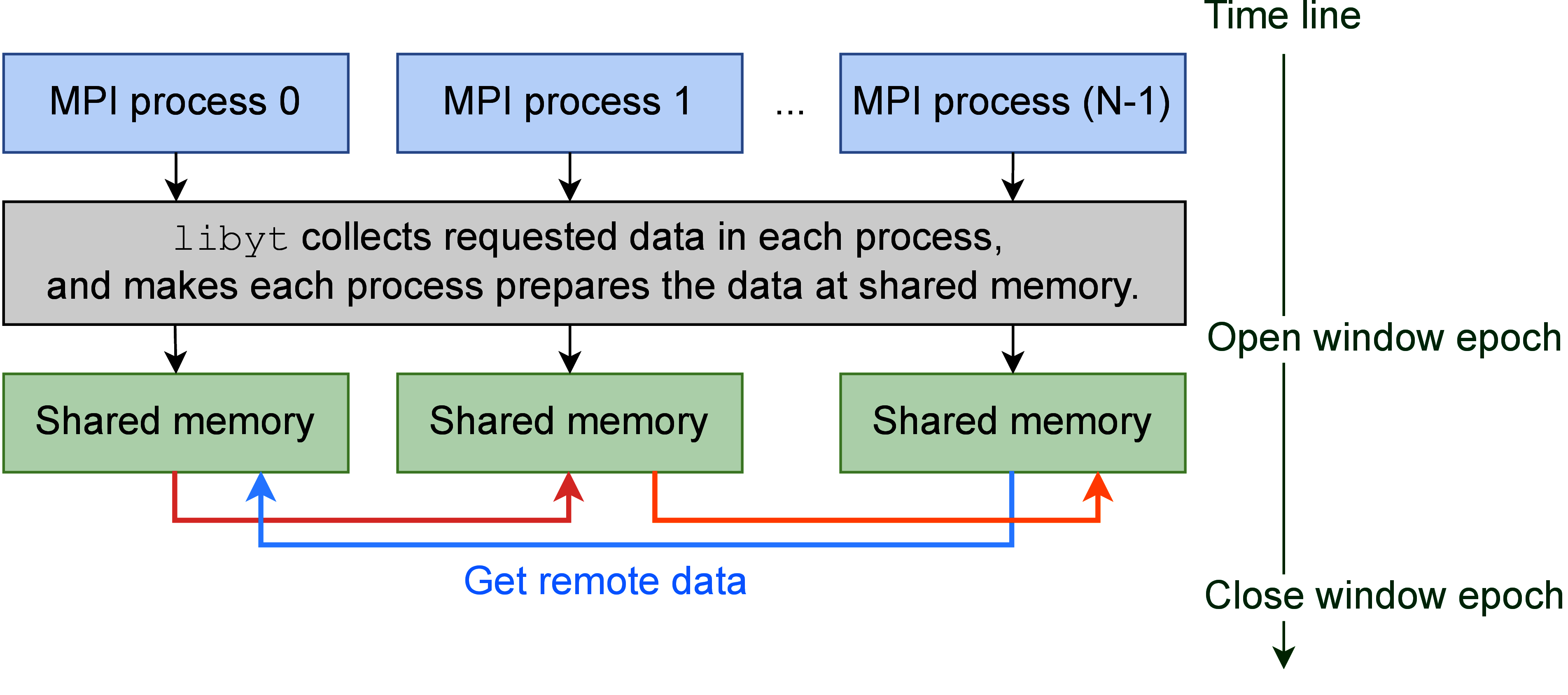
Figure 4:This is the workflow of how libyt redistributes data.
It is done via one-sided communication in MPI.
Each process prepares the requested data from other processes, after this, every process
fetches data located on different processes.
This is a collective operation, and data is redistributed during this window epoch.
Since the data fetched from other processes is only for analysis purpose, it gets freed once Python doesn’t need it at all.
Executing Python Codes and Handling Errors¶
libyt imports user’s Python script at the initialization stage.
Every Python statement is executed inside the imported script’s namespace using PyRun_SimpleString.
The namespace holds Python functions and objects. Every change made will also be stored under this
namespace and will be brought to the following round.
Using libyt for in situ analysis is just like running Python scripts in post-processing.
The only difference lies in how the data is loaded.
Post-processing has everything store on hard disk, while data in in situ analysis is distributed in
memory space in different computing nodes.
Though libyt can call arbitrary Python modules, here, we focus on using yt as the core method.
This is an example of doing slice plot using yt function SlicePlot in post-processing:
1 2 3 4 5 6 7 8 9import yt yt.enable_parallelism() def do_sliceplot(data): ds = yt.load(data) slc = yt.SlicePlot(ds, "z", ("gamer", "Dens")) if yt.is_root(): slc.save() if __name__ == "__main__": do_sliceplot("Data000000")
Converting the post-processing script to inline script is a two-line change.
We need to import yt_libyt[10], which is the yt frontend for libyt.
And then we change yt.load to yt_libyt.libytDataset(). That’s it!
Now data is loaded from libyt instead of loading from hard disk.
The following is the inline Python script:
1 2 3 4 5 6 7 8import yt_libyt import yt yt.enable_parallelism() def do_sliceplot_inline(): ds = yt_libyt.libytDataset() slc = yt.SlicePlot(ds, "z", ("gamer", "Dens")) if yt.is_root(): slc.save()
Simulation can call Python function using libyt API yt_run_Function and
yt_run_FunctionArguments. For example, this calls the Python function do_sliceplot_inline:
yt_run_Function("do_sliceplot_inline");Beside calling Python function, libyt also provides interactive prompt for user to update Python
function, enter statements, and get feedbacks instantly.[11]
This is like running Python prompt inside the ongoing simulation with access to data.
Figure 5 describes the workflow.
The root process takes user inputs and checks the syntax through compiling it to code object using
Py_CompileString. If error occurs, it parses the error to see if this is caused by input not done
yet or a real error.
If it is indeed caused by user hasn’t done yet, for example, when using an if statement,
the prompt will continue waiting for user inputs. Otherwise, it simply prints the error to inform the user.
If the code can be compiled successfully, the root process broadcasts the code to every other MPI
processes.
Then they evaluate the code using PyEval_EvalCode inside the script’s namespace simultaneously.
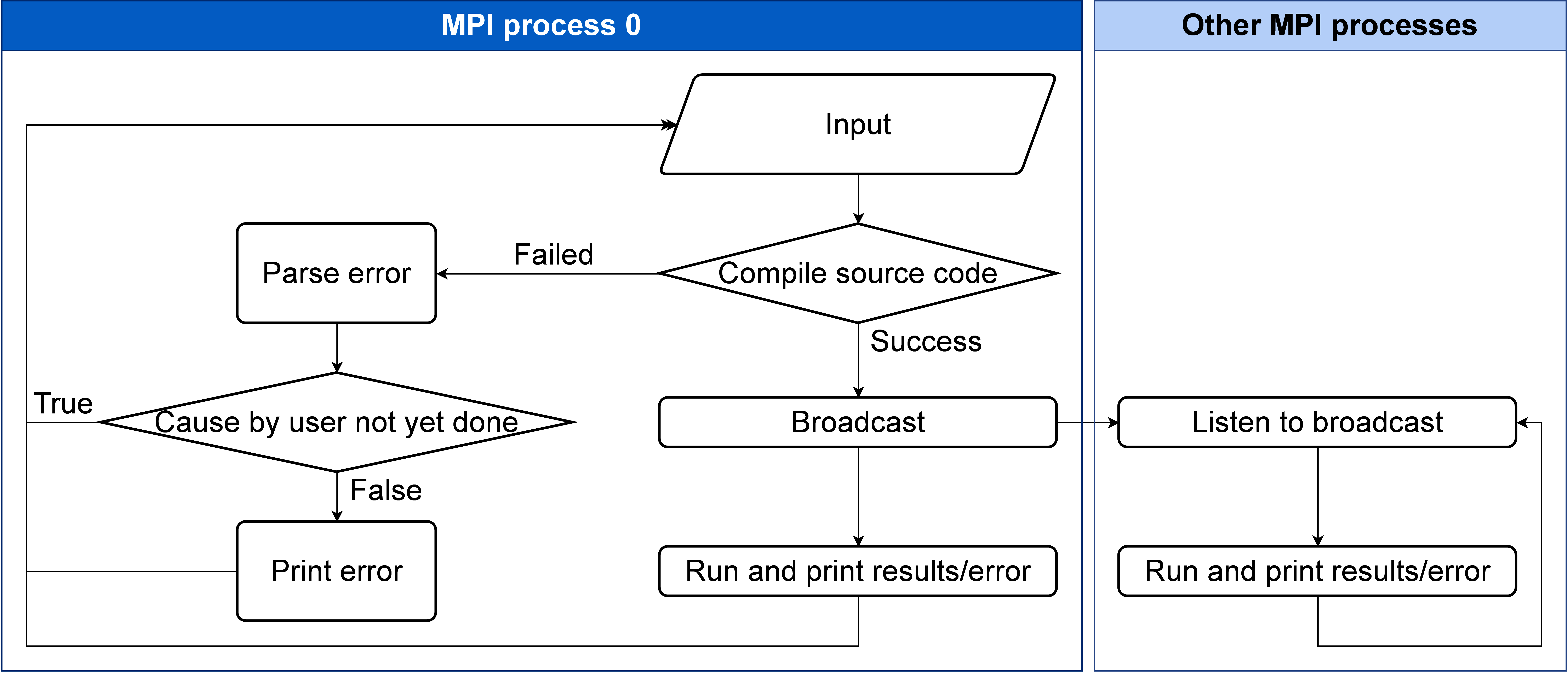
Figure 5:The procedure shows how libyt supports interactive Python prompt.
It takes user inputs on root process and executes Python codes across whole MPI processes.
The root process handles syntax errors and distinguishes whether or not the error is caused
by user hasn’t done inputing yet.
Applications¶
libyt has already been implemented in GAMER[12] Schive et al., 2018 and Enzo[13] Bryan et al., 2014.
GAMER is a GPU-accelerated adaptive mesh refinement code for astrophysics.
It features extremely high performance and parallel scalability and supports a rich set of physics
modules. Enzo is a community-developed adaptive mesh refinement simulation code,
designed for rich, multi-physics hydrodynamic astrophysical calculations.
Here, we demonstrate the results from GAMER using libyt, and we show how libyt solves the
problem of limitation in disk space and improves disk usage efficiency.
Analyzing Fuzzy Dark Matter Vortices Simulation¶
Fuzzy dark matter (FDM) is a promising dark matter candidate Schive et al., 2014.
It is best described by a classical scalar field governed by the Schrödinger-Poisson equation,
because of the large de Broglie wavelength compared to the mean interparticle separation.
FDM halos feature a central compact solitonic core surrounded by fluctuating density granules
resulting from wave function interference.
Quantum vortices can form in density voids caused by fully destructive interference
Chiueh, 1998 Hui et al., 2021.
The dynamics of these vortices in FDM halo have not been investigated thoroughly,
due to the very high spatial and temporal resolution is required, which leads to tremendously
huge disk space. libyt provides a promising approach for this study.
We use GAMER to simulate the evolution of an FDM halo on the Taiwania 3[14].
We use 560 CPU cores by launching 20 MPI processes with 28 OpenMP threads per MPI process to
run the simulation.
The simulation box size is pc, covered by a 6403 base-level grid with
six refinement levels.
The highest level has a maximum resolution of 6.2 pc, so that it is able to resolve the fine
structure and dynamical evolution of vortices within a distance of 3200 pc from the center.
To properly capture the dynamics, we aim for analyzing vortex properties with a temporal resolution of
Myr, corresponding to 321 analysis samples.
Each simulation snapshot, including density, real part, imaginary part, gravitational potential,
and AMR grid information, takes 116 GB.
It will take TB if we do this in post-processing, which is really expensive.
However, it is actually unnecessary to dump all these snapshots since our region of interest is only
the vortex lines around the halo center.
We solve this by using libyt to invoke yt function covering_grid to extract a
uniform-resolution grid centered at the halo center and store these grid data instead of simulation
snapshots on disk.
The uniform grid has dimension 10243 with spatial resolution 6.2 pc (i.e., the
maximum resolution in the simulation), correspnding to the full extracted uniform grid width of
6300 pc.
By storing only the imaginary and real parts of the wave function in single precision,
each sample step now consumes only 8 GB, which is 15 times smaller than the snapshot required in
post-processing.
We further analyze these uniform grids in post-processing, and do volume rendering and create
animation[15] using ParaView Ahrens et al., 2005.
Figure 6 is the volume rendering of the result.
Vortex lines and rings are manifest in the entire domain.
Figure 7 shows a zoom in version of Figure 6, where reconnection of vortex lines take place.
With the help of libyt, we are able to achieve a very high temporal resolution and very high
spatial resolution at the same time.
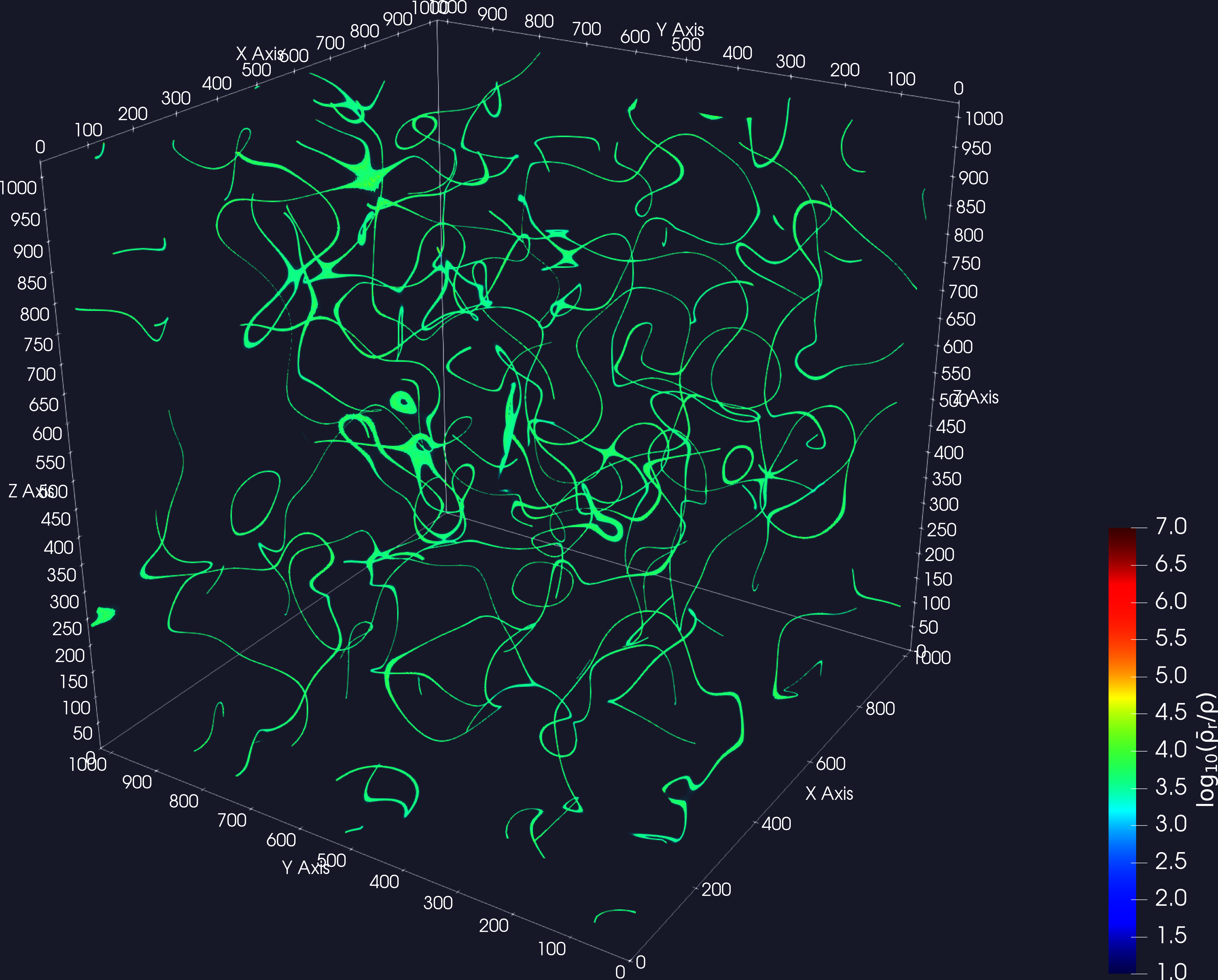
Figure 6:Volume rendering of quantum vortices in a fuzzy dark matter halo with GAMER. Here we use
libyt to extract uniform-resolution data from an AMR simulation on-the-fly, and then visualize
it with ParaView in post-processing. The colormap is the logarithm of reciprocal of density
averaging over radial density profile, which highlights the fluctuations and null density. Tick
labels represent cell indices.
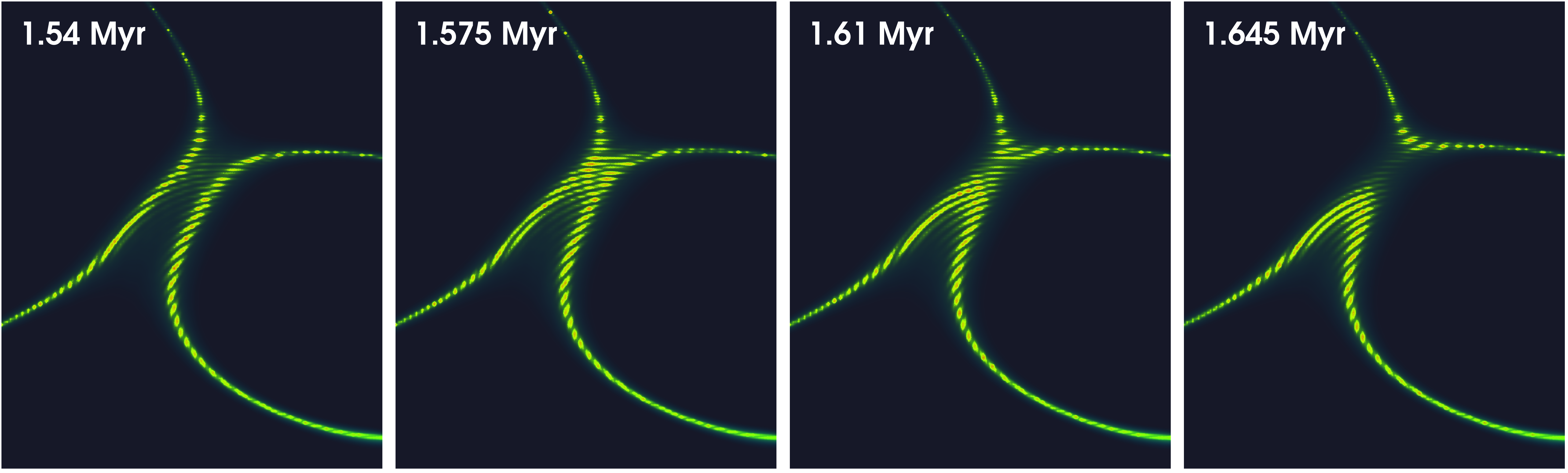
Figure 7:Vortex reconnection process in a fuzzy dark matter halo. This is the result we get if we zoom in to one of the vortex lines in Figure 6 where reconnection of lines take place. We are able to clearly capture the dynamics, and at the same time, preserve high spatial resolution.
Analyzing Core-Collapse Supernova Simulation¶
We use GAMER to simulate core-collapse supernova explosions. The simulations have been
performed on a local cluster using 64 CPU cores and 4 GPUs by launching 8 MPI processes with
8 OpenMP threads per MPI process, and having two MPI processes access the same GPU.
The simulations involve a rich set of physics modules, including
hydrodynamics, self-gravity, a parameterized light-bulb scheme for neutrino
heating and cooling with a fixed neutrino luminosity Couch, 2013,
a parameterized deleptonization scheme Liebendörfer, 2005,
an effective general relativistic potential O'Connor & Couch, 2018, and
a nuclear equation of state Steiner et al., 2013.
For the hydrodynamics scheme, we adopt
the van Leer predictor-corrector integrator Falle, 1991 Leer, 2006,
the piecewise parabolic method for spatial data reconstruction Colella & Woodward, 1984,
and the HLLC Riemann solver Toro, 2009.
The simulation box size is km.
The base-level grid dimension is 1603 and there are eight refinement levels,
reaching a maximum spatial resolution of km.
We use libyt to closely monitor the simulation progress during runtime,
such as the grid refinement distribution, the status and location of shock wave (e.g.,
stalling, revival, breakout), and the evolution of the central proto-neutron star.
libyt calls yt function SlicePlot to draw entropy distribution every
ms. Figure 8 is the output in a time step.
Since entropy is not part of the variable in simulation’s iterative process,
these entropy data will only be generated through user-defined C function,
which in turn calls the nuclear equation of state defined inside GAMER to get entropy,
when they are needed by yt.
libyt tries to minimize memory usage by generating relevant data only.
We can combine every output figure and animate the actual simulation process[16]
without storing any datasets.
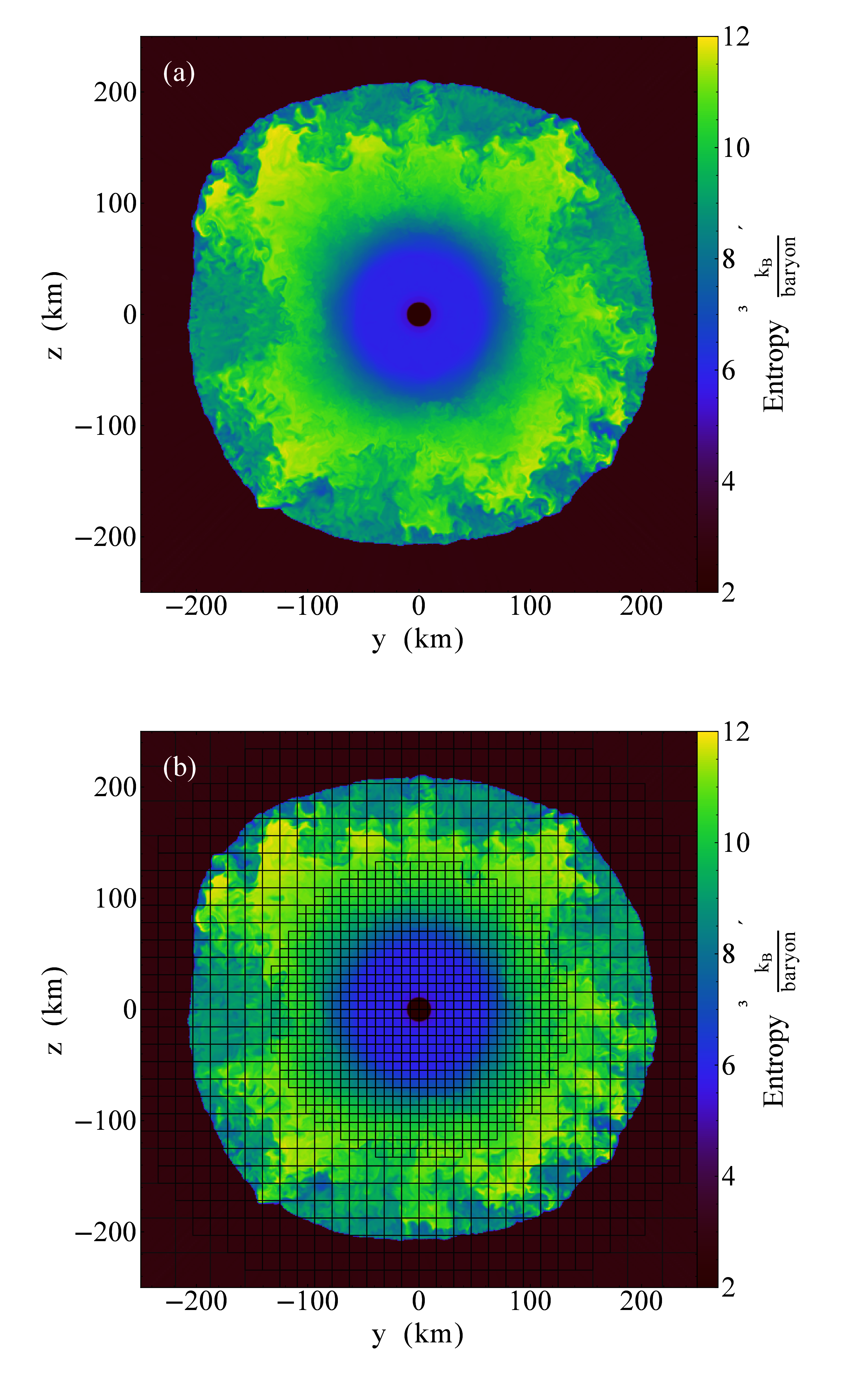
Figure 8:Entropy distribution in a core-collapse supernova simulated by GAMER and plotted by
yt function SlicePlot using libyt.
Plot (a) shows a thin slice cut through the central proto-neutron in the post-bounce phase.
The proto-neutron star has a radius of km and the shock stalls at
km.
Plot (b) shows the underlying AMR grid structure, where each grid consists of 163
cells.
Discussions¶
libyt is free and open source, which does not depend on any non-free or non-open source software.
Converting the post-processing script to inline script is a two-line change, which lowers the barrier
of using this in situ analysis tool.
Though currently, only simulations that use AMR grid data structure are supported by libyt,
we will extend to more data structure (e.g., octree, particle, unstructured mesh, etc) and hope
to engage more simulations and data structures in the future.
Using libyt does not add time penalty to the analysis process, because using Python for in situ analysis
and post-processing are exactly the same, except that the former one reads data from memory and the
latter one reads data from disks. Figure 9 shows the strong scaling of libyt.
The test compares the performance between in situ analysis with libyt and post-processing for
computing 2D profiles on a GAMER dataset.
The dataset contains seven adaptive mesh refinement levels with a total of
cells.
libyt outperforms post-processing by , since it
avoids loading data from disk to memory.
libyt and post-processing have similar deviation from the ideal scaling since libyt directly
borrows the algorithm in yt.
Some improvements have been made in yt, while some are still undergoing to eliminate the scaling
bottleneck.
But also, due to some parts cannot be parallelized, like the import of Python and the current data structure,
the speed up is saturated at large number of processors and can be described by Amdahl’s law.
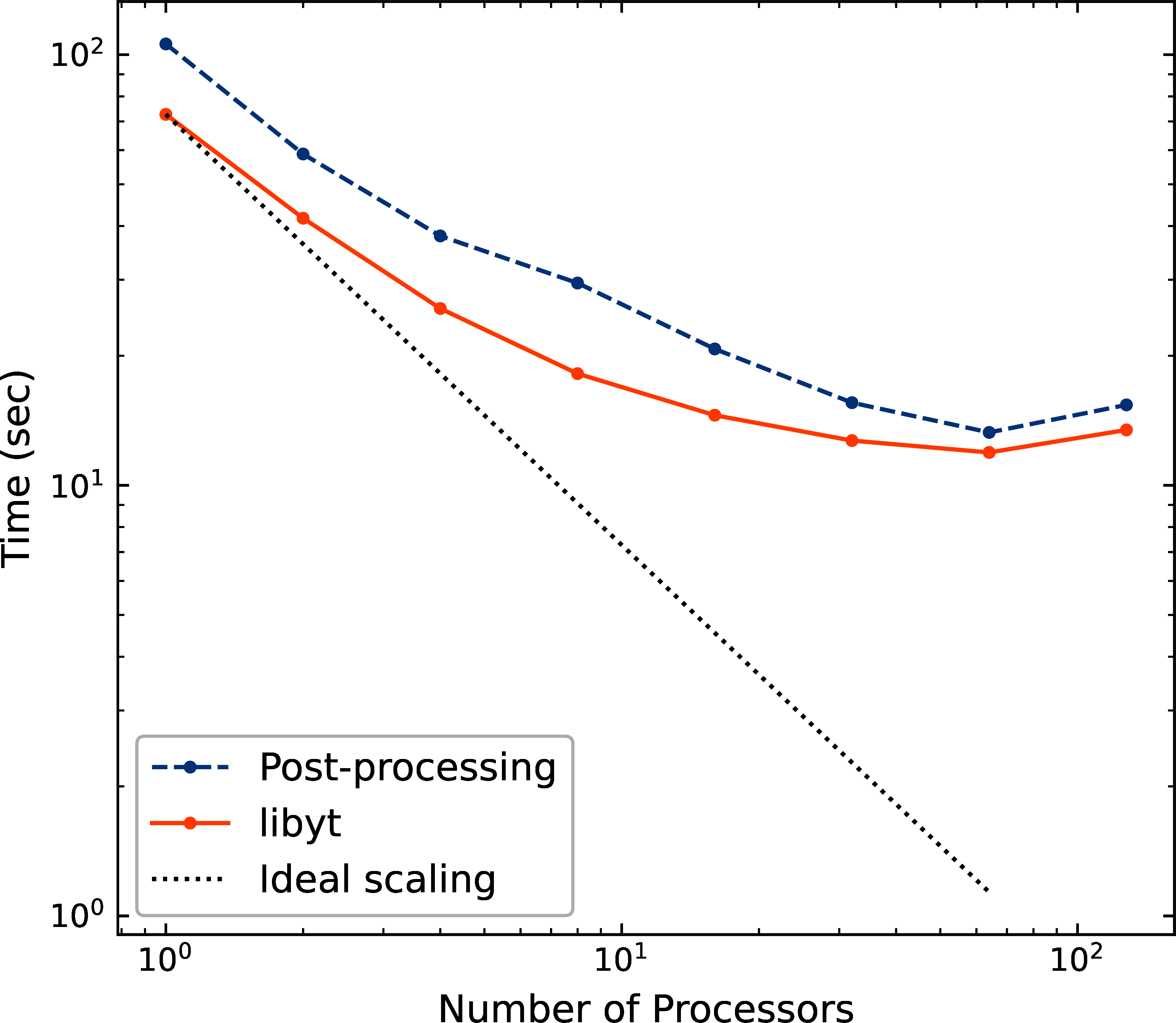
Figure 9:Strong scaling of libyt.
libyt outperforms post-processing by since the former
avoids loading data from disk to memory. The dotted line is the ideal scaling.
libyt and post-processing show a similar deviation from the ideal scaling because it directly
borrows the algorithm in yt. Improvements have been made and will be made in yt to
eliminate the scaling bottleneck.
libyt provides a promising solution that binds simulation to Python with minimal memory overhead
and no additional time penalty.
It makes analyzing large scale simulation feasible, and it can analyze the data with much higher frequency.
It also reduces the barrier of heavy computational jobs written in C/C++ to use Python tools,
which are normally well-developed.
libyt focuses on using yt as its core analytic method, even though it can call other Python
modules, and has the ability to enable back-communication of simulation information.
A use case of this tool could be using yt to select data and then make it as an input source for further analysis.
libyt provides us another way to interact with simulation and data.
Copyright © 2023 Tsai et al. This is an open-access article distributed under the terms of the Creative Commons Attribution 3.0 Unported license.
For more details, please refer to
libytdocuments. (https://yt -project .github .io /libyt /libytAPI) We will support more data structures (e.g., octree, unstructured mesh grid, etc) in the future.
This can be done by using
PyArray_CLEARFLAGSto clear writable flagNPY_ARRAY_WRITEABLE.This can be done by using
PyArray_ENABLEFLAGSto enable own-data flagNPY_ARRAY_OWNDATA.See Parallel Computation With yt for more details.
mpi4pyis Python bindings for MPI. (https://mpi4py .readthedocs .io /en /stable /) ytfunctionalities likefind_max,ProjectionPlot,create_profile,PhasePlot, etc are based on data locality, others likeOffAxisProjectionPlot,SlicePlot,OffAxisSlicePlot, etc don’t.Currently,
libytinteractive prompt only works on local machine or submitting the job to HPC platforms using interactive queue (e.g.,qsub -Ion PBS scheduler). We will support accessing through Jupyter Notebook in the future.Supercomputer at the National Center for High-performance Computing in Taiwan. (https://
www .nchc .org .tw /)
- AMR
- adaptive mesh refinement
- API
- application programming interface
- FDM
- Fuzzy dark matter
- MPI
- message passing interface
- RMA
- Remote Memory Access
- Turk, M. J., Smith, B. D., Oishi, J. S., Skory, S., Skillman, S. W., Abel, T., & Norman, M. L. (2011). yt: A Multi-code Analysis Toolkit for Astrophysical Simulation Data. The Astrophysical Journal Supplement Series, 192, 9. 10.1088/0067-0049/192/1/9
- Harris, C. R., Millman, K. J., van der Walt, S. J., Gommers, R., Virtanen, P., Cournapeau, D., Wieser, E., Taylor, J., Berg, S., Smith, N. J., Kern, R., Picus, M., Hoyer, S., van Kerkwijk, M. H., Brett, M., Haldane, A., del Río, J. F., Wiebe, M., Peterson, P., … Oliphant, T. E. (2020). Array programming with NumPy. Nature, 585(7825), 357–362. 10.1038/s41586-020-2649-2
- Van Rossum, G., & Drake, F. L. (2009). Python 3 Reference Manual. CreateSpace.
- Schive, H.-Y., ZuHone, J. A., Goldbaum, N. J., Turk, M. J., Gaspari, M., & Cheng, C.-Y. (2018). gamer-2: a GPU-accelerated adaptive mesh refinement code – accuracy, performance, and scalability. Monthly Notices of the Royal Astronomical Society, 481(4), 4815–4840. 10.1093/mnras/sty2586
- Bryan, G. L., Norman, M. L., O’Shea, B. W., Abel, T., Wise, J. H., Turk, M. J., Reynolds, D. R., Collins, D. C., Wang, P., Skillman, S. W., Smith, B., Harkness, R. P., Bordner, J., Kim, J. -h., Kuhlen, M., Xu, H., Goldbaum, N., Hummels, C., Kritsuk, A. G., … The Enzo Collaboration. (2014). ENZO: An Adaptive Mesh Refinement Code for Astrophysics. The Astrophysical Journal Supplement Series, 211, 19. 10.1088/0067-0049/211/2/19
- Schive, H.-Y., Chiueh, T., & Broadhurst, T. (2014). Cosmic structure as the quantum interference of a coherent dark wave. Nature Physics, 10, 496–499.
- Chiueh, T. (1998). Dynamical quantum chaos as fluid turbulence. Phys. Rev. E, 57(4), 4150–4154. 10.1103/PhysRevE.57.4150
- Hui, L., Joyce, A., Landry, M. J., & Li, X. (2021). Vortices and waves in light dark matter. Journal of Cosmology and Astroparticle Physics, 2021(1), 011. 10.1088/1475-7516/2021/01/011
- Ahrens, J., Geveci, B., & Law, C. (2005). Visualization Handbook (C. D. Hansen & C. R. Johnson, Eds.; p. 717). Burlington, MA: Elsevier Butterworth–Heinemann.
- Couch, S. M. (2013). The Dependence of the Neutrino Mechanism of Core-collapse Supernovae on the Equation of State. The Astrophysical Journal, 765(1), 29. 10.1088/0004-637X/765/1/29
- Liebendörfer, M. (2005). A Simple Parameterization of the Consequences of Deleptonization for Simulations of Stellar Core Collapse. The Astrophysical Journal, 633(2), 1042–1051. 10.1086/466517
- O’Connor, E. P., & Couch, S. M. (2018). Two-dimensional Core-collapse Supernova Explosions Aided by General Relativity with Multidimensional Neutrino Transport. The Astrophysical Journal, 854(1), 63. 10.3847/1538-4357/aaa893
- Steiner, A. W., Hempel, M., & Fischer, T. (2013). Core-collapse Supernova Equations of State Based on Neutron Star Observations. The Astrophysical Journal, 774(1), 17. 10.1088/0004-637X/774/1/17
- Falle, S. A. E. G. (1991). Self-similar jets. Monthly Notices of the Royal Astronomical Society, 250(3), 581–596. 10.1093/mnras/250.3.581
- van Leer, B. (2006). Upwind and high-resolution methods for compressible flow: From donor cell to residual-distribution schemes. Communications in Computational Physics, 1, 192–206. 10.2514/6.2003-3559
