A Novel Pipeline for Cell Instance Segmentation, Tracking and Motility Classification of Toxoplasma Gondii in 3D Space
Abstract¶
Toxoplasma gondii is the parasitic protozoan that causes disseminated toxoplasmosis, a disease that is estimated to infect around one-third of the world’s population. While the disease is commonly asymptomatic, the success of the parasite is in large part due to its ability to easily spread through nucleated cells. The virulence of T. gondii is predicated on the parasite’s motility. Thus the inspection of motility patterns during its lytic cycle has become a topic of keen interest. Current cell tracking projects usually focus on cell images captured in 2D which are not a true representation of the actual motion of a cell. Current 3D tracking projects lack a comprehensive pipeline covering all phases of preprocessing, cell detection, cell instance segmentation, tracking, and motion classification, and merely implement a subset of the phases. Moreover, current 3D segmentation and tracking pipelines are not targeted for users with less experience in deep learning packages. Our pipeline, TSeg, on the other hand, is developed for segmenting, tracking, and classifying the motility phenotypes of T. gondii in 3D microscopic images. Although TSeg is built initially focusing on T. gondii, it provides generic functions to allow users with similar but distinct applications to use it off-the-shelf. Interacting with all of TSeg’s modules is possible through our Napari plugin which is developed mainly off the familiar SciPy scientific stack. Additionally, our plugin is designed with a user-friendly GUI in Napari which adds several benefits to each step of the pipeline such as visualization and representation in 3D. TSeg proves to fulfill a better generalization, making it capable of delivering accurate results with images of other cell types.
Introduction¶
Quantitative cell research often requires the measurement of different cell properties including size, shape, and motility. This step is facilitated using segmentation of imaged cells. With fluorescent markers, computational tools can be used to complete segmentation and identify cell features and positions over time. 2D measurements of cells can be useful, but the more difficult task of deriving 3D information from cell images is vital for metrics such as motility and volumetric qualities.
Toxoplasmosis is an infection caused by the intracellular parasite Toxoplasma gondii. T. gondii is one of the most successful parasites, infecting at least one-third of the world’s population. Although Toxoplasmosis is generally benign in healthy individuals, the infection has fatal implications in fetuses and immunocompromised individuals Saadatnia & Golkar, 2012. T. gondii’s virulence is directly linked to its lytic cycle which is comprised of invasion, replication, egress, and motility. Studying the motility of T. gondii is crucial in understanding its lytic cycle in order to develop potential treatments.
For this reason, we present a novel pipeline to detect, segment, track, and classify the motility pattern of T. gondii in 3D space. One of the main goals is to make our pipeline intuitively easy to use so that the users who are not experienced in the fields of machine learning (ML), deep learning (DL), or computer vision (CV) can still benefit from it. The other objective is to equip it with the most robust and accurate set of segmentation and detection tools so that the end product has a broad generalization, allowing it to perform well and accurately for various cell types right off the shelf.
PlantSeg uses a variant of 3D U-Net, called Residual 3D U-Net, for preprocessing and segmentation of multiple cell types Wolny et al., 2020. PlantSeg performs best among Deep Learning algorithms for 3D Instance Segmentation and is very robust against image noise Kar et al., 2021. The segmentation module also includes the optional use of CellPose Stringer et al., 2021. CellPose is a generalized segmentation algorithm trained on a wide range of cell types and is the first step toward increased optionality in TSeg. The Cell Tracking module consolidates the cell particles across the z-axis to materialize cells in 3D space and estimates centroids for each cell. The tracking module is also responsible for extracting the trajectories of cells based on the movements of centroids throughout consecutive video frames, which is eventually the input of the motion classifier module.
Most of the state-of-the-art pipelines are restricted to 2D space which is not a true representative of the actual motion of the organism. Many of them require knowledge and expertise in programming, or in machine learning and deep learning models and frameworks, thus limiting the demographic of users that can use them. All of them solely include a subset of the aforementioned modules (i.e. detection, segmentation, tracking, and classification) Stringer et al., 2021. Many pipelines rely on the user to train their own model, hand-tailored for their specific application. This demands high levels of experience and skill in ML/DL and consequently undermines the possibility and feasibility of quickly utilizing an off-the-shelf pipeline and still getting good results.
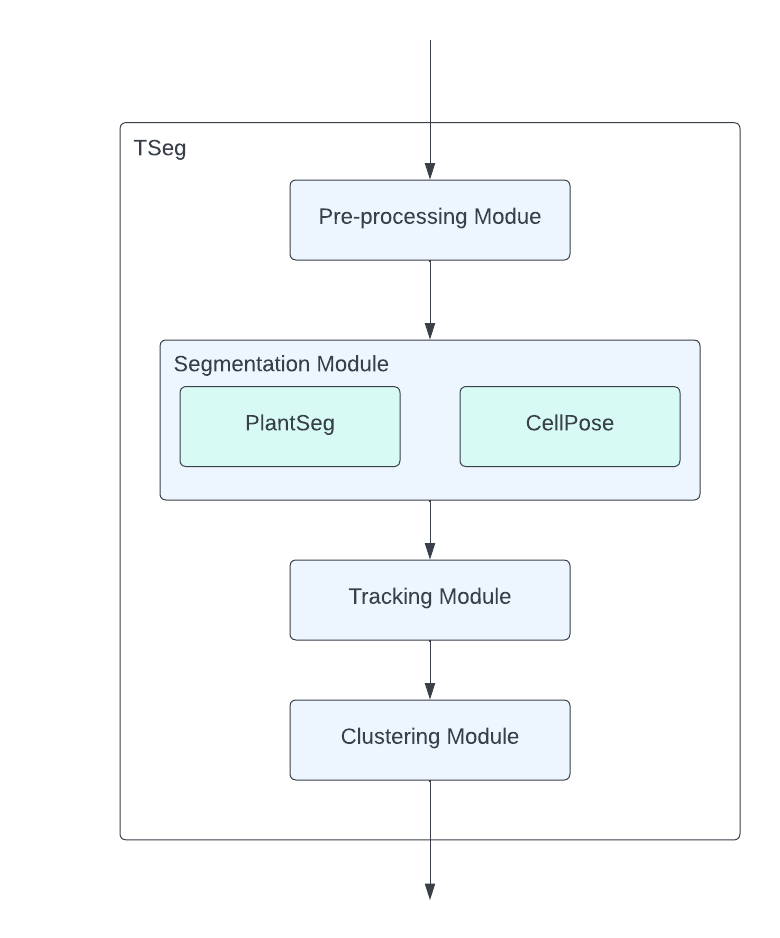
Figure 1:The overview of TSeg’s architecture.
To address these we present TSeg. It segments T. gondii cells in 3D microscopic images, tracks their trajectories, and classifies the motion patterns observed throughout the 3D frames. TSeg is comprised of four modules: pre-processing, segmentation, tracking, and classification. We developed TSeg as a plugin for Napari Sofroniew et al., 2022 - an open-source fast and interactive image viewer for Python designed for browsing, annotating, and analyzing large multi-dimensional images. Having TSeg implemented as a part of Napari not only provides a user-friendly design but also gives more advanced users the possibility to attach and execute their custom code and even interact with the steps of the pipeline if needed. The preprocessing module is equipped with basic and extra filters and functionalities to aid in the preparation of the input data. TSeg gives its users the advantage of utilizing the functionalities that PlantSeg and CellPose provide. These functionalities can be chosen in the pre-processing, detection, and segmentation steps. This brings forth a huge variety of algorithms and pre-built models to select from, making TSeg not only a great fit for T. gindii, but also a variety of different cell types.
The rest of this paper is structured as follows: After briefly reviewing the literature in Related Work, we move on to thoroughly describe the details of our work in the Method section. Following that, the Results section depicts the results of comprehensive tests of our plugin on T. gondii cells.
Related Work¶
The recent solutions in generalized and automated segmentation tools are focused on 2D cell images. Segmentation of cellular structures in 2D is important but not representative of realistic environments. Microbiological organisms are free to move on the z-axis and tracking without taking this factor into account cannot guarantee a full representation of the actual motility patterns. As an example, Fazli et al. (2018) identified three distinct motility types for T. gondii with two-dimensional data, however, they also acknowledge and state that based established heuristics from previous works there are more than three motility phenotypes for T. gondii. The focus on 2D research is understandable due to several factors. 3D data is difficult to capture as tools for capturing 3D slices and the computational requirements for analyzing this data are not available in most research labs. Most segmentation tools are unable to track objects in 3D space as the assignment of related centroids is more difficult. The additional noise from capture and focus increases the probability of incorrect assignment. 3D data also has issues with overlapping features and increased computation required per frame of time.
Fazli et al. (2018) studies the motility patterns of T. gondii and provides a computational pipeline for identifying motility phenotypes of T. gondii in an unsupervised, data-driven way. In that work Ca2+ is added to T. gondii cells inside a Fetal Bovine Serum. T. gondii cells react to Ca2+ and become motile and fluorescent. The images of motile T. gondii cells were captured using an LSM 710 confocal microscope. They use Python 3 and associated scientific computing libraries (NumPy, SciPy, scikit-learn, matplotlib) in their pipeline to track and cluster the trajectories of T. gondii. Based on this work Fazli et al. (2018) work on another pipeline consisting of preprocessing, sparsification, cell detection, and cell tracking modules to track T. gondii in 3D video microscopy where each frame of the video consists of image slices taken 1 micro-meters of focal depth apart along the z-axis direction. In their latest work Fazli et al. (2019) developed a lightweight and scalable pipeline using task distribution and parallelism. Their pipeline consists of multiple modules: reprocessing, sparsification, cell detection, cell tracking, trajectories extraction, parametrization of the trajectories, and clustering. They could classify three distinct motion patterns in T. gondii using the same data from their previous work.
While combining open source tools is not a novel architecture, little has been done to integrate 3D cell tracking tools. Fazeli et al. (2020) motivated by the same interest in providing better tools to non-software professionals created a 2D cell tracking pipeline. This pipeline combines Stardist Weigert et al., 2020 and TrackMate Tinevez et al., 2017 for automated cell tracking. This pipeline begins with the user loading cell images and centroid approximations to the ZeroCostDL4Mic Chamier et al., 2021 platform. ZeroCostDL4Mic is a deep learning training tool for those with no coding expertise. Once the platform is trained and masks for the training set are made for hand-drawn annotations, the training set can be input to Stardist. Stardist performs automated object detection using Euclidean distance to probabilistically determine cell pixels versus background pixels. Lastly, Trackmate uses segmentation images to track labels between timeframes and display analytics.
This Stardist pipeline is similar in concept to TSeg. Both create an automated segmentation and tracking pipeline but TSeg is oriented to 3D data. Cells move in 3-dimensional space that is not represented in a flat plane. TSeg also does not require the manual training necessary for the other pipeline. Individuals with low technical expertise should not be expected to create masks for training or even understand the training of deep neural networks. Lastly, this pipeline does not account for imperfect datasets without the need for preprocessing. All implemented algorithms in TSeg account for microscopy images with some amount of noise.
Wen et al. (2021) combines multiple existing new technologies including deep learning and presents 3DeeCellTracker. 3DeeCellTracker segments and tracks cells on 3D time-lapse images. Using a small subset of their dataset they train the deep learning architecture 3D U-Net for segmentation. For tracking, a combination of two strategies was used to increase accuracy: local cell region strategies, and spatial pattern strategy. Kapoor & Carabaña (2021) presents VollSeg that uses deep learning methods to segment, track, and analyze cells in 3D with irregular shape and intensity distribution. It is a Jupyter Notebook-based Python package and also has a UI in Napari. For tracking, a custom tracking code is developed based on Trackmate.
Many segmentation tools require some amount of knowledge in Machine or Deep Learning concepts. Training the neural network in creating masks is a common step for open-source segmentation tools. Automating this process makes the pipeline more accessible to microbiology researchers.
Method¶
Data¶
Our dataset consists of 11 videos of T. gondii cells under a microscope, obtained from different experiments with different numbers of cells. The videos are on average around 63 frames in length. Each frame has a stack of 41 image slices of size 500×502 pixels along the z-axis (z-slices). The z-slices are captured 1µm apart in optical focal length making them 402µm×401µm×40µm in volume. The slices were recorded in raw format as RGB TIF images but are converted to grayscale for our purpose. This data is captured using a PlanApo 20x objective (NA = 0:75) on a preheated Nikon Eclipse TE300 epifluorescence microscope. The image stacks were captured using an iXon 885 EMCCD camera (Andor Technology, Belfast, Ireland) cooled to -70°C and driven by NIS Elements software (Nikon Instruments, Melville, NY) as part of related research by Leung et al. (2014). The camera was set to frame transfer sensor mode, with a vertical pixel shift speed of 1:0 µs, vertical clock voltage amplitude of +1, readout speed of 35MHz, conversion gain of 3:8×, EM gain setting of 3 and 22 binning, and the z-slices were imaged with an exposure time of 16ms.
Software¶
Napari Plugin¶
TSeg is developed as a plugin for Napari - a fast and interactive multi-dimensional image viewer for python that allows volumetric viewing of 3D images Sofroniew et al., 2022. Plugins enable developers to customize and extend the functionality of Napari. For every module of TSeg, we developed its corresponding widget in the GUI, plus a widget for file management. The widgets have self-explanatory interface elements with tooltips to guide the inexperienced user to traverse through the pipeline with ease. Layers in Napari are the basic viewable objects that can be shown in the Napari viewer. Seven different layer types are supported in Napari: Image, Labels, Points, Shapes, Surface, Tracks, and Vectors, each of which corresponds to a different data type, visualization, and interactivity Sofroniew et al., 2022. After its execution, the viewable output of each widget gets added to the layers. This allows the user to evaluate and modify the parameters of the widget to get the best results before continuing to the next widget. Napari supports bidirectional communication between the viewer and the Python kernel and has a built-in console that allows users to control all the features of the viewer programmatically. This adds more flexibility and customizability to TSeg for the advanced user. The full code of TSeg is available on GitHub under the MIT open source license at https://
Computational Pipeline¶
Pre-Processing¶
Due to the fast imaging speed in data acquisition, the image slices will inherently have a vignetting artifact, meaning that the corners of the images will be slightly darker than the center of the image. To eliminate this artifact we added adaptive thresholding and logarithmic correction to the pre-processing module. Furthermore, another prevalent artifact on our dataset images was a Film-Grain noise (AKA salt and pepper noise). To remove or reduce such noise a simple gaussian blur filter and a sharpening filter are included.
Cell Detection and Segmentation¶
TSeg’s Detection and Segmentation modules are in fact backed by PlantSeg and CellPose. The Detection Module is built only based on PlantSeg’s CNN Detection Module Wolny et al., 2020 , and for the Segmentation Module, only one of the three tools can be selected to be executed as the segmentation tool in the pipeline. Naturally, each of the tools demands specific interface elements different from the others since each accepts different input values and various parameters. TSeg orchestrates this and makes sure the arguments and parameters are passed to the corresponding selected segmentation tool properly and the execution will be handled accordingly. The parameters include but are not limited to input data location, output directory, and desired segmentation algorithm. This allows the end-user complete control over the process and feedback from each step of the process. The preprocessed images and relevant parameters are sent to a modular segmentation controller script. As an effort to allow future development on TSeg, the segmentation controller script shows how the pipeline integrates two completely different segmentation packages. While both PlantSeg and CellPose use conda environments, PlantSeg requires modification of a YAML file for initialization while CellPose initializes directly from command line parameters. In order to implement PlantSeg, TSeg generates a YAML file based on GUI input elements. After parameters are aligned, the conda environment for the chosen segmentation algorithm is opened in a subprocess. The $CONDA_PREFIX environment variable allows the bash command to start conda and context switch to the correct segmentation environment.
Tracking¶
Features in each segmented image are found using the scipy label function. In order to reduce any leftover noise, any features under a minimum size are filtered out and considered leftover noise. After feature extraction, centroids are calculated using the center of mass function in scipy. The centroid of the 3D cell can be used as a representation of the entire body during tracking. The tracking algorithm goes through each captured time instance and connects centroids to the likely next movement of the cell. Tracking involves a series of measures in order to avoid incorrect assignments. An incorrect assignment could lead to inaccurate result sets and unrealistic motility patterns. If the same number of features in each frame of time could be guaranteed from segmentation, minimum distance could assign features rather accurately. Since this is not a guarantee, the Hungarian algorithm must be used to associate a COST with the assignment of feature tracking. The Hungarian method is a combinatorial optimization algorithm that solves the assignment problem in polynomial time. COST for the tracking algorithm determines which feature is the next iteration of the cell’s tracking through the complete time series. The combination of distance between centroids for all previous points and the distance to the potential new centroid. If an optimal next centroid can’t be found within an acceptable distance of the current point, the tracking for the cell is considered as complete. Likewise, if a feature is not assigned to a current centroid, this feature is considered a new object and is tracked as the algorithm progresses. The complete path for each feature is then stored for motility analysis.
Motion Classification¶
To classify the motility pattern of T. gondii in 3D space in an unsupervised fashion we implement and use the method that Fazli et al. (2019) introduced. In that work, they used an autoregressive model (AR); a linear dynamical system that encodes a Markov-based transition prediction method. The reason is that although K-means is a favorable clustering algorithm, there are a few drawbacks to it and to the conventional methods that draw them impractical. Firstly, K-means assumes Euclidian distance, but AR motion parameters are geodesics that do not reside in a Euclidean space, and secondly, K-means assumes isotropic clusters, however, although AR motion parameters may exhibit isotropy in their space, without a proper distance metric, this issue cannot be clearly examined Fazli et al., 2019.
Conclusion and Discussion¶
TSeg is an easy to use pipeline designed to study the motility patterns of T. gondii in 3D space. It is developed as a plugin for Napari and is equipped with a variety of deep learning based segmentation tools borrowed from PlantSeg and CellPose, making it a suitable off-the-shelf tool for applications incorporating images of cell types not limited to T. gondii. Future work on TSeg includes the expansion of implemented algorithms and tools in its preprocessing, segmentation, tracking, and clustering modules.
Copyright © 2022 Vaezi et al. This is an open-access article distributed under the terms of the Creative Commons Attribution 3.0 Unported license.
- AR
- autoregressive model
- CV
- computer vision
- DL
- deep learning
- ML
- machine learning
- Saadatnia, G., & Golkar, M. (2012). A review on human toxoplasmosis. Scandinavian Journal of Infectious Diseases, 44(11), 805–814. 10.3109/00365548.2012.693197
- Wolny, A., Cerrone, L., Vijayan, A., Tofanelli, R., Barro, A. V., Louveaux, M., Wenzl, C., Strauss, S., Wilson-Sánchez, D., Lymbouridou, R., Steigleder, S. S., Pape, C., Bailoni, A., Duran-Nebreda, S., Bassel, G. W., Lohmann, J. U., Tsiantis, M., Hamprecht, F. A., Schneitz, K., … Kreshuk, A. (2020). Accurate and versatile 3D segmentation of plant tissues at cellular resolution. eLife, 9, e57613. 10.7554/eLife.57613
- Kar, A., Petit, M., Refahi, Y., Cerutti, G., Godin, C., & Traas, J. (2021). Assessment of deep learning algorithms for 3D instance segmentation of confocal image datasets. bioRxiv. 10.1101/2021.06.09.447748
- Stringer, C., Wang, T., Michaelos, M., & Pachitariu, M. (2021). Cellpose: a generalist algorithm for cellular segmentation. Nature Methods, 18(1), 100–106. 10.1101/2020.02.02.931238
- Sofroniew, N., Lambert, T., Evans, K., Nunez-Iglesias, J., Bokota, G., Winston, P., Peña-Castellanos, G., Yamauchi, K., Bussonnier, M., Doncila Pop, D., Can Solak, A., Liu, Z., Wadhwa, P., Burt, A., Buckley, G., Sweet, A., Migas, L., Hilsenstein, V., Gaifas, L., … McGovern, A. (2022). napari: a multi-dimensional image viewer for Python (v0.4.16) [Computer software]. Zenodo. 10.5281/zenodo.6598542
- Fazli, M. S., Velia, S. A., Moreno, S. N., & Quinn, S. (2018). Unsupervised discovery of toxoplasma gondii motility phenotypes. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), 981–984. 10.1109/isbi.2018.8363735
- Fazli, M. S., Vella, S. A., Moreno, S. N., Ward, G. E., & Quinn, S. P. (2018). Toward simple & scalable 3D cell tracking. 2018 IEEE International Conference on Big Data (Big Data), 3217–3225. 10.1109/BigData.2018.8622403
- Fazli, M. S., Stadler, R. V., Alaila, B., Vella, S. A., Moreno, S. N., Ward, G. E., & Quinn, S. (2019). Lightweight and scalable particle tracking and motion clustering of 3D cell trajectories. 2019 IEEE International Conference on Data Science and Advanced Analytics (DSAA), 412–421. 10.1109/dsaa.2019.00056
- Fazeli, E., Roy, N. H., Follain, G., Laine, R. F., von Chamier, L., Hänninen, P. E., Eriksson, J. E., Tinevez, J.-Y., & Jacquemet, G. (2020). Automated cell tracking using StarDist and TrackMate. F1000Research, 9. 10.12688/f1000research.27019.1
- Weigert, M., Schmidt, U., Haase, R., Sugawara, K., & Myers, G. (2020, March). Star-convex Polyhedra for 3D Object Detection and Segmentation in Microscopy. 2020 IEEE Winter Conference on Applications of Computer Vision (WACV). 10.1109/wacv45572.2020.9093435
- Tinevez, J.-Y., Perry, N., Schindelin, J., Hoopes, G. M., Reynolds, G. D., Laplantine, E., Bednarek, S. Y., Shorte, S. L., & Eliceiri, K. W. (2017). TrackMate: An open and extensible platform for single-particle tracking. Methods, 115, 80–90. https://doi.org/10.1016/j.ymeth.2016.09.016
- von Chamier, L., Laine, R. F., Jukkala, J., Spahn, C., Krentzel, D., Nehme, E., Lerche, M., Hernández-Pérez, S., Mattila, P. K., Karinou, E., & others. (2021). Democratising deep learning for microscopy with ZeroCostDL4Mic. Nature Communications, 12(1), 1–18. 10.1038/s41467-021-22518-0
- Wen, C., Miura, T., Voleti, V., Yamaguchi, K., Tsutsumi, M., Yamamoto, K., Otomo, K., Fujie, Y., Teramoto, T., Ishihara, T., Aoki, K., Nemoto, T., Hillman, E. M., & Kimura, K. D. (2021). 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images. Elife, 10. 10.7554/eLife.59187
- Kapoor, V., & Carabaña, C. (2021). Cell Tracking in 3D using deep learning segmentations. Python in Science Conference, 154–161. 10.25080/majora-1b6fd038-014
- Leung, J., Rould, M., Konradt, C., Hunter, C., & Ward, G. (2014). Disruption of TgPHIL1 Alters Specific Parameters of Toxoplasma gondii Motility Measured in a Quantitative, Three-Dimensional Live Motility Assay. PloS One, 9, e85763. 10.1371/journal.pone.0085763
