
SciPy Proceedings: An Exemplar for Publishing Computational Open Science
Abstract¶
The SciPy Proceedings have served as a cornerstone of scholarly communication within the scientific Python ecosystem since their introduction in 2008. In 2024, the publication process underwent a significant transformation, adopting a new open-source infrastructure built on MyST Markdown and Curvenote. This transition enabled a web-first, interactive publishing workflow that improves reproducibility, readability, and metadata quality. Authors submit articles through GitHub, receive new structured feedback via continuous integration (CI), and benefit from new live previews, automated PDF generation with Typst, and archival Journal Article Tag Suite (JATS) XML export. This paper details the infrastructure, authoring workflow, and technical tooling that powers the new SciPy Proceedings. We demonstrate how these advances support executable content, interactive figures, and modern metadata standards—offering a model for computational publishing in other scientific communities.
1Introduction¶
Scientific publishing faces a fundamental disconnect between published research and the computational methods that produced it. While researchers increasingly rely on code, data, and interactive computational workflows, traditional academic publishing remains rooted in static, paper-based formats that cannot capture the dynamic nature of modern scientific inquiry. This is particularly acute in computational science, where reproducibility requires access to executable code, interactive visualizations, and the ability to verify results through direct experimentation.
The reproducibility crisis in science has been well-documented, with studies showing that a significant portion of published research cannot be replicated due to missing code, data, or insufficient methodological detail American Association for the Advancement of Science (AAAS), 2015Baker, 2016. In computational research, this problem is amplified by the inherent complexity of software-based methods, where subtle differences in environments, dependencies, or implementation details can lead to dramatically different results Stodden et al., 2016. Traditional PDF-based publishing, while excellent for narrative content, fundamentally cannot execute code, provide interactive exploration of data, or adapt to different computational environments.
Since its inception in 2008, the SciPy Proceedings have offered a high-quality venue for publishing scientific and technical contributions built on the Python ecosystem. The SciPy Proceedings are an opportunity for accepted speakers at the SciPy Conference to submit a peer-reviewed publication, and there have been over 350 peer-reviewed articles published. The articles often represent a chance to document or understand the motivations behind new scientific software or tools built for the scientific Python community. This type of work is critical for advancement across many research disciplines, but traditional academic journals can be reluctant to publish it. Without the SciPy Proceedings, many of these articles may not exist.
In 2024, the SciPy Proceedings committee reimagined the entire publishing workflow by adopting modern, open-source community-built infrastructure built on MyST Markdown and Curvenote. This transition enabled the SciPy Proceedings to be part of a larger movement and ecosystem of open source publishing tools and workflows. The result was a new model of computational publishing that supports executable content, high-quality structured metadata, and a web-first reading experience with interactive features. This shift not only enhances the publishing experience for authors and reviewers but also significantly improves the way readers interact with scientific content, fundamentally advancing the principles of open science and reproducible research.

Figure 1:Side-by-side comparison of a 2023 Proceedings listing page, which only included titles, authors, and links to PDFs, and the 2024 Proceedings listing in the new format, using modern web components to render thumbnails and additional metadata.
2A New Model for Scientific Publishing¶
The most significant improvement in the 2024 Proceedings is the transition from static PDFs to web-native articles rendered directly from MyST Markdown (https://mystmd.org). Each article is hosted by Curvenote on the SciPy Proceedings website as an interactive web page, preserving semantic structure and supporting accessibility, deep linking, and citation preview. This transition addresses the fundamental limitations of traditional publishing by enabling the kind of interactive, executable content that computational research requires.
2.1Executable Content and Reproducibility¶
The new format enables true reproducibility by allowing readers to execute code directly within the article. Articles can include executed Jupyter Notebooks (e.g. Lujan (2024)) with full output from code cells already present, interactive code blocks that can be re-run, live data connections that pull from versioned repositories, and environment specifications that document the exact computational environment used. This executable content transforms the reading experience from passive consumption to see the computational work directly integrated into the reading experience. This direct engagement builds trust in the research and enables the kind of iterative exploration that drives scientific discovery.
2.2Enhanced Discoverability and Navigation¶
The web-native format also provides discoverability through structured content and semantic markup. Articles feature linkable sections, figures, and code blocks that enable precise citation and reference (e.g. to a specific section or figure), semantic structure that search engines can understand and index, rich metadata that connects articles to related research, datasets, and software, and cross-references that link to related work, dependencies, and extensions.
2.3Interactive Exploration and Visualization¶
The new format supports the kind of interactive exploration that computational research demands: 3D interactive visualizations using tools like k3d, pythreejs, or vtk.js that respond to user input, dynamic plots that update based on parameter changes, embedded computational widgets that allow real-time experimentation, and responsive design that adapts to different devices and screen sizes. These interactive elements transform static figures into dynamic tools for understanding. Readers can explore data from multiple angles, test hypotheses through direct manipulation, and gain deeper insights through hands-on exploration.

Figure 2:We have focused on web-native rendering, hover previews and including interactive figures. Showing content from McCormick & Elliott (2024) and Cockett et al. (2024).
2.4The Authoring Workflow¶
The new authoring and submission workflow is aimed at transforming the traditional publishing process into a collaborative, iterative experience that leverages modern software development practices. Authors write their articles using a standardized MyST template available on GitHub from SciPy. Each submission consists of a myst.yml file capturing comprehensive article metadata, including authorship and author Open Researcher and Contributor IDs (ORCIDs), affiliations and Research Organization Registry IDs (RORs), keywords, and licensing; Markdown or Jupyter Notebook source files using MyST syntax, including the main article and any supplementary material; and static assets such as figures and datasets, with clear organization and documentation.
After previewing the article locally with myst start, authors open a Pull Request on GitHub. This submission triggers a GitHub Actions Continuous Integration (CI) pipeline that performs comprehensive quality checks: structural validation of the document to ensure proper formatting and organization, metadata schema validation (e.g., required ORCID, DOIs for references) to ensure completeness, content validation including word count limits, figure references, and citation completeness, rendering to HTML for preview to catch formatting issues early, PDF generation via Typst to ensure print compatibility, and Journal Article Tag Suite (JATS) XML export for archival and indexing.
This automated quality assurance process serves multiple purposes. It catches common errors early in the submission process (See Figure 3), reducing the burden on reviewers and authors. It ensures consistency across all articles in the proceedings, improving the overall quality of the publication. It provides immediate feedback to authors, enabling rapid iteration and improvement—with authors getting structured and actionable feedback in less than a minute.
The GitHub-based workflow, popularized by the Journal of Open Source Software (JOSS) Smith et al., 2018Diehl et al., 2025, enables a collaborative review that is more transparent and effective than traditional peer review processes. Reviewers often leave line-level comments directly on the source content, enabling precise feedback, suggest changes through GitHub’s suggestion feature, making it easy for authors to accept improvements, track the evolution of articles through the commit history, understanding how feedback was incorporated, and collaborate with other reviewers through threaded discussions, building consensus on improvements.
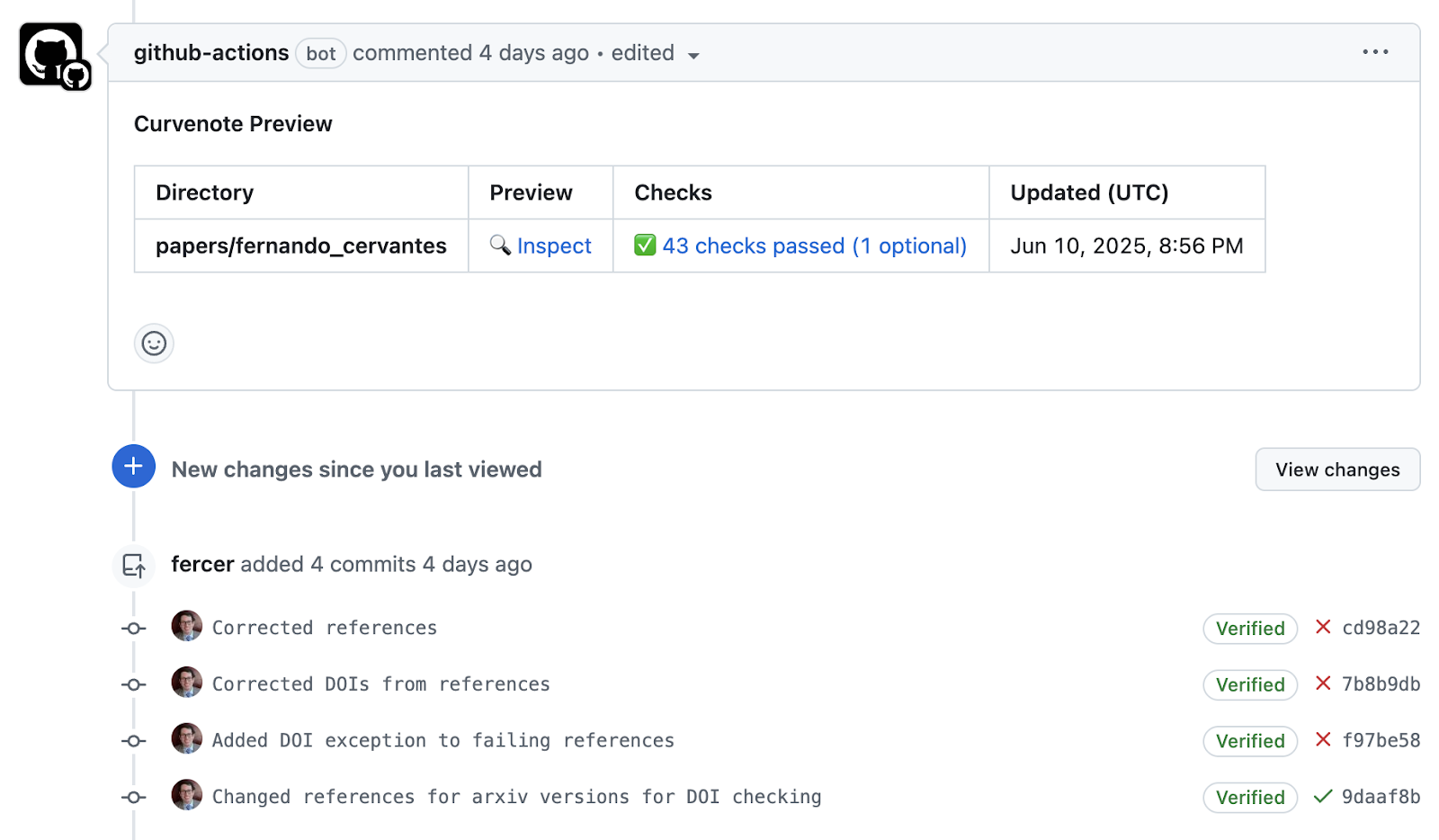
Figure 3:Screenshot of a GitHub Pull Request showing CI failure due to missing ORCID metadata, with the corrected version in a follow-up commit (Cervantes-Sanchez, 2025).
3Technical Foundation¶
The transition to the new SciPy Proceedings infrastructure required consideration of tool selection to balance functionality, openness, accessibility, and community needs. Each component was chosen to advance the goals of reproducible, interactive scientific publishing while maintaining the open-source ethos of the SciPy community.
The markup language that the proceedings used for 16 years was reStructuredText and a restricted version of LaTeX was added in 2022. In 2024, MyST Markdown was selected as the core authoring format for several reasons. As an extension of standard Markdown, it provides a familiar, accessible authoring experience while adding the semantic structure needed for scientific publishing. MyST supports executable content through Jupyter integration, rich metadata through YAML frontmatter, cross-references and citations, mathematical notation through LaTeX syntax, and multiple output formats including HTML, PDF, and JATS XML. The tooling also supports LaTeX directly, so the options introduced in 2022 were still supported. MyST is developed as an open-source project with strong community involvement, ensuring that the format evolves with the needs of scientific publishing while remaining accessible to all contributors; the project is governed under Project Jupyter.
The Curvenote CLI is based on top of MyST and is also available under MIT license, it adds specific metadata checks that can be run locally as well as through the open source GitHub actions. The PDF template is SciPy-specific and can be used for any MyST content or forked for other proceedings. The Curvenote CLI can be used to send a draft of a submission to web hosting infrastructure, managed by Curvenote, to display the built manuscripts and publish them; the Curvenote platform is commercial and enables publishing management and the hosting of in-progress drafts. For institutions or individuals who prefer to self-host this style of proceedings, the open-source components can combined independently using MyST Markdown directly (see MyST documentation for deployment instructions), providing full control over the end-to-end infrastructure. Examples of this include Neurolibre, a journal that hosts interactive preprints and has a similar vision for interactive scientific narratives Karakuzu, 2025; Neurolibre use very similar open-source infrastructure, including MyST Markdown and Open Journals, which powers JOSS.
The transition from LaTeX to Typst represents a significant modernization of the PDF generation process. LaTeX, while powerful, is complex, fragile and has large dependency chains. Build times can be unpredictable, and subtle changes in document structure can cause complex compilation failures. Typst offers significantly faster compilation times and more predictable behavior, reducing the friction in the authoring and review process. Typst provides a more modern, programmable approach to typesetting that enables the creation of highly customized templates. This allowed us to build a SciPy-specific template that maintains consistency across all articles while supporting the advanced formatting requirements of scientific publishing (See Figure 4). Typst is developed as an open-source project, ensuring that the typesetting engine remains accessible and modifiable by the community.
The transition to Typst has delivered measurable improvements: PDF generation is now 5-10x faster than with LaTeX, compilation failures are rare and easily diagnosed, the SciPy template can be easily forked, modified or extended, and it supports advanced typography and layout features (See Figure 4).
The full archive of scientific content over the 18 years has been transitioned to an open GitHub archive that includes the built assets (JATS, PDF, and Manuscript Exchange Common Approach (MECA) files) for every paper; these were previously available through each branch of the SciPy Proceedings repository.
3.1Build System and Output Formats¶
The SciPy Proceedings infrastructure is built on Curvenote’s GitHub Actions and the Curvenote CLI, both available through the MIT License. Upon opening a submission pull request and on each subsequent commit, content is automatically validated. This validation ensures all content is formatted correctly so the article can fully build. It also checks that all required metadata is in place and all submission guidelines, such as word count, number of keywords, etc. are met. After this validation the submission is rendered across several formats using a reproducible build pipeline:
Web (HTML): Rendered using MyST Markdown and published to the Proceedings site.
PDF: Generated using Typst, replacing LaTeX for greater speed, reliability, and customizability.
JATS XML: Introduced in 2024, this machine-readable format supports long-term preservation and interoperability with scholarly indexing systems.
Previews: The previews from the Pull Request are hosted by Curvenote, allowing reviewers and authors to inspect the rendered output, exactly as it will appear in the final proceedings publication[1].
Figure 4:Comparison between 2023 and 2024 PDF templates, 2024 improved the branding and template to be more modern; the template now uses Typst over LaTeX, which also improves the build speed. The template is available under the MIT license.
The transition to Typst allowed us to build a fully customized SciPy Proceedings PDF template, ensuring consistency with the new web style while supporting traditional academic formatting and citation standards.
3.2Metadata and Interoperability¶
The move to structured MyST Markdown and schema-validated metadata unlocks new possibilities for reuse, attribution, and discoverability. Metadata is required at the submission stage and validated throughout the review process, including author names and ORCIDs, institutional affiliations (RORs), keywords, license, and abstract, and DOI for citations and associated datasets or software.
Each component of an article (e.g., figures, equations, or code blocks) is preserved in a structured way, improving reusability. For example, figures can be referenced externally, cited independently, or embedded into institutional repositories without modification. JATS XML export supports interoperability with discovery engines and indexing services, while also laying the foundation for cross-journal content federation and machine-readable citation networks.
As more conferences, journals, preprint servers, and MyST-powered documentation sites adopt the structured information and metadata standards, these resources create an interconnected network of scientific knowledge that is browsable, inter-linked, and discoverable. Each article becomes a node in this network, with rich metadata, cross-references, and executable content that can be linked, cited, and built upon from any project. This can allow for reuse from other projects, for example:
Referencing figures from other publications, Lujan, 2024;
Pulling in context via hover previews from Cockett et al., 2024;
Showing rich interactive visualizations from McCormick & Elliott, 2024; or
Showing videos from Bao et al., 2024.
These cross-references provide direct access to a modular component of a research article or resource and can be shown in context (try hovering over one of the links above in the web-version, or watch the video in Figure 5). These are standard features in any MyST Markdown powered document Cockett et al., 2024, and provide exciting possibilities for new in-context reading experiences.
Figure 5:Hover cross-references between articles, using the MyST metadata and content standards.
3.3Demonstrating Accessibility¶
The SciPy Proceedings model provides a replicable template for other conferences and journals seeking to modernize their publishing practices. The modular architecture and open-source foundation enable easy adoption by other scientific communities. The accessibility of this publishing model is perhaps best demonstrated by Morganton Scientific, a high school science journal that publishes using the same infrastructure as the SciPy Proceedings Curvenote Inc., 2024. The fact that high school students can successfully publish scientific content using the same tools and workflows as professional researchers demonstrates how intuitive and accessible this approach is. This accessibility is crucial for democratizing scientific publishing and ensuring that valuable contributions from diverse communities can be shared and recognized.
The success of Morganton Scientific shows that the barrier to entry for modern scientific publishing can be remarkably low when the right infrastructure is in place. This has implications for educational institutions, citizen science projects, and emerging research communities that might otherwise struggle to establish traditional publishing channels.
4Conclusion¶
As computational research continues to grow in importance across all scientific disciplines, the need for publishing infrastructure that can effectively communicate this work will only increase. The SciPy Proceedings demonstrate that this challenge can be met through community-driven development, open-source tools, and a commitment to the fundamental principles of scientific communication. The 2024 transition addresses the core challenges of reproducibility, interactivity, and discoverability in scientific publishing. Through modern web technologies and open-source infrastructure, the Proceedings have demonstrated that scientific communication can evolve to meet the needs of modern research while maintaining the rigor and quality that the scientific community expects.
The new SciPy Proceedings infrastructure provides a working example of how scientific publishing can embrace the interactive, executable nature of computational research. By combining MyST Markdown, Curvenote, GitHub-native review workflows, and Typst, the Proceedings offer authors and readers a more dynamic, structured, and metadata-rich experience for computational sciences.
The infrastructure is designed to be replicable by other scientific communities. The modular architecture, open-source components, and documented processes provide a template that can be adapted for different domains and use cases. The success of the SciPy Proceedings demonstrates that this approach can work at scale and provides a path for other communities seeking to modernize their publishing practices.
We gratefully acknowledge the contributions of Chris Calloway, Meghann Agarwal, Stefan van der Walt, and Jim Weis for their support, coordination, and feedback throughout the transition to the new SciPy Proceedings infrastructure. Their efforts were instrumental in shaping the submission process, reviewing tooling, and ensuring a smooth experience for authors and reviewers alike.
We thank the current SciPy 2025 Proceedings Committee — Amey Ambade, Ana Comesana, Sean Freeman, Sanhita Joshi, Charles Lindsey, and Hongsup Shin — for their dedication and hard work in supporting authors, managing reviews, and ensuring the quality and integrity of this year’s volume.
We also thank the many members of past SciPy Proceedings committees and infrastructure teams whose work laid the foundation for this transformation. The continuity, care, and community leadership built over the last 17 years have made this evolution possible.
Copyright © 2025 Cockett et al. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license, which enables reusers to distribute, remix, adapt, and build upon the material in any medium or format, so long as attribution is given to the creators.
For fully open-source workflows, these previews can be hosted by GitHub pages or similar static rendering sites (see GitHub Pages deployment).
- CI
- Continuous Integration
- CLI
- Command Line Interface
- DOI
- Digital Object Identifier
- HTML
- Hypertext Markup Language
- ID
- Identifier
- JATS
- Journal Article Tag Suite
- JOSS
- Journal of Open Source Software
- MyST
- Markedly Structured Text
- ORCID
- Open Researcher and Contributor ID
- Portable Document Format
- ROR
- Research Organization Registry
- XML
- Extensible Markup Language
- YAML
- Yet Another Markup Language
- Estimating the reproducibility of psychological science. (2015). Science, 349(6251). 10.1126/science.aac4716
- Baker, M. (2016). 1,500 scientists lift the lid on reproducibility. Nature, 533(7604), 452–454. 10.1038/533452a
- Stodden, V., McNutt, M., Bailey, D. H., Deelman, E., Gil, Y., Hanson, B., Heroux, M. A., Ioannidis, J. P. A., & Taufer, M. (2016). Enhancing reproducibility for computational methods. Science, 354(6317), 1240–1241. 10.1126/science.aah6168
- Lujan, A. (2024). multinterp: A Unified Interface for Multivariate Interpolation in the Scientific Python Ecosystem. Proceedings of the 23rd Python in Science Conference, 1–19. 10.25080/fgcj9164
- McCormick, M., & Elliott, P. (2024). ITK-Wasm: Universal spatial analysis and visualization. Proceedings of the 23rd Python in Science Conference, 256–279. 10.25080/tcfj5130
- Cockett, R., Purves, S., Koch, F., & Morrison, M. (2024). Continuous Tools for Scientific Publishing: Using MyST Markdown and Curvenote to encourage continuous science practices. Proceedings of the 23rd Python in Science Conference, 121–136. 10.25080/nkvc9349
- Smith, A. M., Niemeyer, K. E., Katz, D. S., Barba, L. A., Githinji, G., Gymrek, M., Huff, K. D., Madan, C. R., Cabunoc Mayes, A., Moerman, K. M., Prins, P., Ram, K., Rokem, A., Teal, T. K., Valls Guimera, R., & Vanderplas, J. T. (2018). Journal of Open Source Software (JOSS): design and first-year review. PeerJ Computer Science, 4, e147. 10.7717/peerj-cs.147
- Diehl, P., Soneson, C., Kurchin, R. C., Mounce, R., & Katz, D. S. (2025). The Journal of Open Source Software (JOSS): Bringing Open-Source Software Practices to the Scholarly Publishing Communityfor Authors, Reviewers, Editors, and Publishers. Journal of Librarianship and Scholarly Communication, 12(2). 10.31274/jlsc.18285
- Karakuzu, A. (2025). Toward a woven literature: Open-source infrastructure for reproducible publishing. 10.55458/neurolibre.00041
- Bao, F. S., Qi, M., Tu, R., & Wan, E. (2024). Funix - The laziest way to build GUI apps in Python: Make it (the app) before they fake it (in Figma or on a napkin). Proceedings of the 23rd Python in Science Conference, 174–195. 10.25080/jfyn3740
- (2024). Morganton Scientific, 2. 10.62329/dwgm3685
