Design of a Scientific Data Analysis Support Platform
Abstract¶
Software data analytic workflows are a critical aspect of modern scientific research and play a crucial role in testing scientific hypotheses. A typical scientific data analysis life cycle in a research project must include several steps that may not be fundamental to testing the hypothesis, but are essential for reproducibility. This includes tasks that have analogs to software engineering practices such as versioning code, sharing code among research team members, maintaining a structured codebase, and tracking associated resources such as software environments. Tasks unique to scientific research include designing, implementing, and modifying code that tests a hypothesis. This work refers to this code as an experiment, which is defined as a software analog to physical experiments.
A software experiment manager should support tracking and reproducing individual experiment runs, organizing and presenting results, and storing and reloading intermediate data on long-running computations. A software experiment manager with these features would reduce the time a researcher spends on tedious busywork and would enable more effective collaboration. This work discusses the necessary design features in more depth, some of the existing software packages that support this workflow, and a custom developed open-source solution to address these needs.
This manuscript has been authored by UT-Battelle, LLC, under
contract DE-AC05-00OR22725 with the US Department of Energy (DOE). The US
government retains and the publisher, by accepting the article for
publication, acknowledges that the US government retains a nonexclusive,
paid-up, irrevocable, worldwide license to publish or reproduce the
published form of this manuscript, or allow others to do so, for US
government purposes. DOE will provide public access to these results of
federally sponsored research in accordance with the DOE Public Access Plan
(http://
Introduction¶
Modern science increasingly uses software as a tool for conducting research and scientific data analyses. The growing number of libraries and frameworks facilitating this work has greatly lowered the barrier to usage, allowing more researchers to benefit from this paradigm. However, as a result of the dependence on software, there is a need for more thorough integration of sound software engineering practices with the scientific process. The fragility of complex environments containing heavily interconnected packages coupled with a lack of provenance of the artifacts generated throughout the development of an experiment increases the potential for long-term problems, undetected bugs, and failure to reproduce previous analyses.
Fundamentally, science revolves around the ability for others to repeat and reproduce prior published works, and this has become a difficult task with many computation-based studies. Often, scientists outside of a computer science field may not have training in software engineering best practices, or they may simply disregard them because the focus of a researcher is on scientific publications rather than the analysis software itself. Lack of documentation and provenance of research artifacts and frequent failure to publish repositories for data and source code has led to a crisis in reproducibility in artificial intelligence (AI) and other fields that rely heavily on computation Stodden et al., 2013Donoho et al., 2009Hutson, 2018. One study showed that quantifiably few machine learning (ML) papers document specifics in how they ran their experiments Gundersen et al., 2018. This gap between established practices from the software engineering field and how computational research is conducted has been studied for some time, and the problems that can stem from it are discussed at length in Storer (2018).
To mitigate these issues, computation-based research requires better infrastructure and tooling Peng, 2011 as well as applying relevant software engineering principles Storer, 2018Dubois, 2005 to allow data scientists to ensure their work is effective, correct, and reproducible. In this paper we focus on the ability to manage reproducible workflows for scientific experiments and data analyses. We discuss the features that software to support this might require, compare some of the existing tools that address them, and finally present the open-source tool Curifactory which incorporates the proposed design elements.
Related Work¶
Reproducibility of AI experiments has been separated into three different degrees Gundersen & Kjensmo, 2018: Experiment reproduciblity, or repeatability, refers to using the same code implementation with the same data to obtain the same results. Data reproducibility, or replicability, is when a different implementation with the same data outputs the same results. Finally, method reproducibility describes when a different implementation with different data is able to achieve consistent results. These degrees are discussed in Gundersen et al. (2018), comparing the implications and trade-offs on the amount of work for the original researcher versus an external researcher, and the degree of generality afforded by a reproduced implementation. A repeatable experiment places the greatest burden on the original researcher, requiring the full codebase and experiment to be sufficiently documented and published so that a peer is able to correctly repeat it. At the other end of the spectrum, method reproducibility demands the greatest burden on the external researcher, as they must implement and run the experiment from scratch. For the remainder of this paper, we refer to “reproducibility” as experiment reproducibility (repeatability). Tooling that is able to assist with documentation and organization of a published experiment reduces the amount of work for the original researcher and still allows for the lowest level of burden to external researchers to verify and extend previous work.
In an effort to encourage better reproducibility based on datasets, the Findable, Accessible, Interoperable, and Reusable (FAIR) data principles Wilkinson et al., 2016 were established. These principles recommend that data should have unique and persistent identifiers, use common standards, and provide rich metadata description and provenance, allowing both humans and machines to effectively parse them. These principles have been extended more broadly to software Lamprecht et al., 2020, computational workflows Goble et al., 2020, and to entire data pipelines Mitchell et al., 2021.
Various works have surveyed software engineering practices and identified practices that provide value in scientific computing contexts, including various forms of unit and regression testing, proper source control usage, formal verification, bug tracking, and agile development methods Storer, 2018Dubois, 2005. In particular, Storer (2018) described many concepts from agile development as being well suited to an experimental context, where the current knowledge and goals may be fairly dynamic throughout the project. They noted that although many of these techniques could be directly applied, some required adaptation to make sense in the scientific software domain.
Similar to this paper, two other works Deelman et al., 2009Wratten et al., 2021 discuss sets of design aspects and features that a workflow manager would need. Deelman et al. describe the life cycle of a workflow as composition, mapping, execution, and provenance capture Deelman et al., 2009. A workflow manager must then support each of these aspects. Composition is how the workflow is constructed, such as through a graphical interface or with a text configuration file. Mapping and execution are determining the resources to be used for a workflow and then utilizing those resources to run it, including distributing to cloud compute and external representational state transfer (REST) services. This also refers to scheduling subworkflows/tasks to reuse intermediate artifacts as available. Provenance, which is crucial for enabling repeatability, is how all artifacts, library versions, and other relevant metadata are tracked during the execution of a workflow.
Wratten, Wilm, and Göke surveyed many bioinformatics pipline and workflow management tools, listing the challenges that tooling should address: data provenance, portability, scalability, and re-entrancy Wratten et al., 2021. Provenance is defined the same way as in Deelman et al. (2009), and further states the need for generating reports that include the tracking information and metadata for the associated experiment run. Portability - allowing set up and execution of an experiment in a different environment - can be a challenge because of the dependency requirements of a given system and the ease with which the environment can be specified and reinitialized on a different machine or operating system. Scalability is important especially when large scale data, many compute-heavy steps, or both are involved throughout the workflow. Scalability in a manager involves allowing execution on a high-performance computing (HPC) system or with some form of parallel compute. Finally they mention re-entrancy, or the ability to resume execution of a compute step from where it last stopped, preventing unnecessary recomputation of prior steps.
One area of the literature that needs further discussion is the design of automated provenance tracking systems. Existing workflow management tools generally require source code modifications to take full advantage of all features. This can entail a significant learning curve and places additional burden on the researcher. To address this, some sources propose automatic documentation of experiments and code through static source code analysis Namaki et al., 2020Redyuk, 2019.
Beyond the preexisting body of knowledge about software engineering principles, other works Sandve et al., 2013Kelly et al., 2009 describe recommended rules and practices to follow when conducting computation-based research. These include avoiding manual data manipulation in favor of scripted changes, keeping detailed records of how results are produced (manual provenance), tracking the versions of libraries and programs used, and tracking random seeds. Many of these ideas can be assisted or encapsulated through appropriate infrastructure decisions, which is the premise on which this work bases its software reviews.
Although this paper focuses on the scientific workflow, a growing related field tackles many of the same issues from an industry standpoint: machine learning operations (MLOps) Goyal, 2020. MLOps, an ML-oriented version of DevOps, is concerned with supporting an entire data science life cycle, from data acquisition to deployment of a production model. Many of the same challenges are present, reproducibility and provenance are crucial in both production and research workflows Ruf et al., 2021. Infrastructure, tools, and practices developed for MLOps may also hold value in the scientific community.
A taxonomy for ML tools that we reference throughout this work is from Quaranta et al. (2021), which describes a characterization of tools consisting of three primary categories: general, analysis support, and reproducibility support, each of which is further subdivided into aspects to describe a tool. For example, these subaspects include data visualization, web dashboard capabilities, experiment logging, and the interaction modes the tool supports, such as a command line interface (CLI) or application programming interface (API).
Design Features¶
We combine the two sets of capabilities from Deelman et al. (2009) and Wratten et al. (2021) with the taxonomy from Quaranta et al. (2021) to propose a set of six design features that are important for an experiment manager. These include orchestration, parameterization, caching, reproducibility, reporting, and scalability. The crossover between these proposed feature sets are shown in Table 1. We expand on each of these in more depth in the subsections below.
Table 1:Comparing design features listed in various works.
| This work | Deelman et al. (2009) | Wratten et al. (2021) | Taxonomy Quaranta et al., 2021 |
|---|---|---|---|
| Orchestration | Composition | --- | Reproducibility/pipeline creation |
| Parameterization | --- | --- | --- |
| Caching | --- | Re-entrancy | --- |
| Reproducibility | Provenance | Provenance, portability | Reproducibility |
| Reporting | --- | --- | Analysis/visualization, web dashboard |
| Scalability | Mapping, execution | Scalability | Analysis/computational resources |
Orchestration¶
Orchestration of an experiment refers to the mechanisms used to chain and compose a sequence of smaller logical steps into an overarching pipeline. This provides a higher-level view of an experiment and helps abstract away some of the implementation details. Operation of most workflow managers is based on a directed acyclic graph (DAG), which specifies the stages/steps as nodes and the edges connecting them as their respective inputs and outputs. The intent with orchestration is to encourage designing distinct, reusable steps that can easily be composed in different ways to support testing different hypotheses or overarching experiment runs. This allows greater focus on the design of the experiments than the implementation of the underlying functions that the experiments consist of. As discussed in the taxonomy Quaranta et al., 2021, pipeline creation can consist of a combination of scripts, configuration files, or a visual tool. This aspect falls within the composition capability discussed in Deelman et al. (2009).
Parameterization¶
Parameterization specifies how a compute pipeline is customized for a particular run by passing in configuration values to change aspects of the experiment. The ability to customize analysis code is crucial to conducting a compute-based experiment, providing a mechanism to manipulate a variable under test to verify or reject a hypothesis.
Conventionally, parameterization is done either through specifying parameters in a CLI call or by passing configuration files in a format like JSON or YAML. As discussed in Deelman et al. (2009), parameterization sometimes consists of more complicated needs, such as conducting parameter sweeps or grid searches. There are libraries dedicated to managing parameter searches like this, such as hyperopt Bergstra et al., 2013 used in Ruf et al. (2021).
Although not provided as a design capability in the other works, we claim the mechanisms provided for parameterization are important, as these mechanisms are the primary way to configure, modify, and vary experiment execution without explicitly changing the code itself or modifying hard-coded values. This means that a recorded parameter set can better “describe” an experiment run, increasing provenance and making it easier for another researcher to understand what pieces of an experiment can be readily changed and explored.
Some support is provided for this in Deelman et al. (2009), stating that the necessity of running many slight variations on workflows sometimes leads to the creation of ad hoc scripts to generate the variants, which leads to increased complexity in the organization of the codebase. Improved mechanisms to parameterize the same workflow for many variants helps to manage this complexity.
Caching¶
Refining experiment code and finding bugs is often a lengthy iterative process, and removing the friction of constantly rerunning all intermediate steps every time an experiment is wrong can improve efficiency. Caching values between each step of an experiment allows execution to resume at a certain spot in the pipeline, rather than starting from scratch every time. This is defined as re-entrancy in Wratten et al. (2021).
In addition to increasing the speed of rerunning experiments and running new experiments that combine old results for analysis, caching is useful to help find and debug mistakes throughout an experiment. Cached outputs from each step allow manual interrogation outside of the experiment. For example, if a cleaning step was implemented incorrectly and a user noticed an invalid value in an output data table, they could use a notebook to load and manipulate the intermediate artifact tables for that data to determine what stage introduced the error and what code should be used to correctly fix it.
Reproducibility¶
Mechanisms for reproducibility are one of the most important features for a successful data analysis support platform. Reproducibility is challenging because of the complexity of constantly evolving codebases, complicated and changing dependency graphs, and inconsistent hardware and environments. Reproducibility entails two subcomponents: provenance and portability. This falls under the provenance aspect from Deelman et al. (2009), both data provenance and portability from Wratten et al. (2021), and the entire reproducibility support section of the taxonomy Quaranta et al., 2021.
Data provenance is about tracking the history, configuration, and steps taken to produce an intermediate or final data artifact. In ML this would include the cleaning/munging steps used and the intermediate tables created in the process, but provenance can apply more broadly to any type of artifact an experiment may produce, such as ML models themselves, or “model provenance” Sugimura & Hartl, 2018. Applying provenance beyond just data is critical, as models may be sensitive to the specific sets of training data and conditions used to produce them Hutson, 2018. This means that everything required to directly and exactly reproduce a given artifact is recorded, such as the manipulations applied to its predecessors and all hyperparameters used within those manipulations.
Portability refers to the ability to take an experiment and execute it outside of the initial computing environment it was created in Wratten et al. (2021). This can be a challenge if all software dependency versions are not strictly defined, or when some dependencies may not be available in all environments. Minimally, allowing portability requires keeping explicit track of all packages and the versions used. A 2017 study Olorisade et al., 2017 found that even this minimal step is rarely taken. Another mechanism to support portability is the use of containerization, such as with Docker or Podman Sugimura & Hartl, 2018.
Reporting¶
Reporting is an important step for analyzing the results of an experiment, through visualizations, summaries, comparisons of results, or combinations thereof. As a design capability, reporting refers to the mechanisms available for the system to export or retrieve these results for human analysis. Although data visualization and analysis can be done manually by the scientist, tools to assist with making these steps easier and to keep results organized are valuable from a project management standpoint. Mechanisms for this might include a web interface for exploring individual or multiple runs. Under the taxonomy Quaranta et al., 2021, this falls primarily within analysis support, such as data visualization or a web dashboard.
Scalability¶
Many data analytic problems require large amounts of space and compute resources, often beyond what can be handled on an individual machine. To efficiently support running a large experiment, mechanisms for scaling execution are important and could include anything from supporting parallel computation on an experiment or stage level, to allowing the execution of jobs on remote machines or within an HPC context. This falls within both mapping and execution from Deelman et al. (2009), the scalability aspect from Wratten et al. (2021), and the computational resources category within the taxonomy Quaranta et al., 2021.
Existing Tools¶
A wide range of pipeline and workflow tools have been developed to support many of these design features, and some of the more common examples include DVC Kuprieiev et al., 2022 and MLFlow MLflow: A Machine Learning Lifecycle Platform, 2022. We briefly survey and analyze a small sample of these tools to demonstrate the diversity of ideas and their applicability in different situations. Table 2 compares the support of each design feature by each tool.
Table 2:Supported design features in each tool. Note, + indicates that a feature is supported, ++ indicates very strong support, and * indicates tooling that supports caching artifacts as a provenance tool but does not provide a mechanism for automatically reloading cached values as a form of re-entrancy.
| Orchestration | Parameterization | Caching | Provenance | Portability | Reporting | Scalability | |
| DVC | + | + | ++ | + | + | + | + |
| MLFlow | + | * | ++ | ++ | ++ | ++ | |
| Sacred | + | ++ | * | ++ | + | + | |
| Kedro | + | + | * | + | ++ | ++ | ++ |
| Curifactory | + | ++ | ++ | ++ | ++ | + | + |
DVC¶
DVC Kuprieiev et al., 2022 is a Git-like version control tool for datasets. Orchestration is done by specifying stages, or runnable script commands, either in YAML or directly on the CLI. A stage is specified with output file paths and input file paths as dependencies, allowing an implicit pipeline or DAG to form, representing all the processing steps. Parameterization is done by defining within a YAML file what the possible parameters are, along with the default values. When running the DAG, parameters can be customized on the CLI. Since inputs and outputs are file paths, caching and re-entrancy come for free, and DVC will intelligently determine if certain stages do not need to be re-computed.
A saved experiment or state is frozen into each commit, so all parameters and artifacts are available at any point. No explicit tracking of the environment (e.g., software versions and hardware info) is present, but this could be manually included by tracking it in a separate file. Reporting can be done by specifying per-stage metrics to track in the YAML configuration. The CLI includes a way to generate HTML files on the fly to render requested plots. There is also an external “Iterative Studio” project, which provides a live web dashboard to view continually updating HTML reports from DVC. For scalability, parallel runs can be achieved by queuing an experiment multiple times in the CLI.
MLFlow¶
MLFlow MLflow: A Machine Learning Lifecycle Platform, 2022 is a framework for managing the entire life cycle of an ML project, with an emphasis on scalability and deployment. It has no specific mechanisms for orchestration, instead allowing the user to intersperse MLFlow API calls in an existing codebase. Runnable scripts can be provided as entry points into a configuration YAML, along with the parameters that can be provided to it. Parameters are changed through the CLI. Although MLFlow has extensive capabilities for tracking artifacts, there are no automatic re-entrancy methods. Reproducibility is a strong feature, and provenance and portability are well supported. The tracking module provides provenance by recording metadata such as the Git commit, parameters, metrics, and any user-specified artifacts in the code. Portability is done by allowing the environment for an entry point to be specified as a Conda environment or Docker container. MLFlow then ensures that the environment is set up and active before running. The CLI even allows directly specifying a GitHub link to an mlflow-enabled project to download, set up, and then run the associated experiment. For reporting, the MLFlow tracking UI lets the user view and compare various runs and their associated artifacts through a web dashboard. For scalability, both distributed storage for saving/loading artifacts as well as execution of runs on distributed clusters is supported.
Sacred¶
Sacred Greff et al., 2017 is a Python library and CLI tool to help organize and reproduce experiments. Orchestration is managed through the use of Python decorators, a “main” for experiment entry point functions and “capture” for parameterizable functions, where function arguments are automatically populated from the active configuration when called. Parameterization is done directly in Python through applying a config decorator to a function that assigns variables. Configurations can also be written to or read from JSON and YAML files, so parameters must be simple types. Different observers can be specified to automatically track much of the metadata, environment information, and current parameters, and within the code the user can specify additional artifacts and resources to track during the run. Each run will store the requested outputs, although there is no re-entrant use of these cached values. Portability is supported through the ability to print the versions of libraries needed to run a particular experiment. Reporting can be done through a specific type of observer, and the user can provide custom templated reports that are generated at the end of each run.
Kedro¶
Kedro Alam et al., 2022 is another Python library/CLI tool for managing reproducible and modular experiments. Orchestration is particularly well done with “node” and “pipeline” abstractions, a node referring to a single compute step with defined inputs and outputs, and a pipeline implemented as an ordered list of nodes. Pipelines can be composed and joined to create an overarching workflow. Possible parameters are defined in a YAML file and either set in other parameter files or configured on the CLI. Similar to MLFlow, while tracking outputs are cached, there’s no automatic mechanism for re-entrancy. Provenance is achieved by storing user-specified metrics and tracked datasets for each run, and it has a few different mechanisms for portability. This includes the ability to export an entire project into a Docker container. A separate Kedro-Viz tool provides a web dashboard to show a map of experiments, as well as showing each tracked experiment run and allowing comparison of metrics and outputs between them. Projects can be deployed into several different cloud providers, such as Databricks and Dask clusters, allowing for several options for scalability.
Curifactory¶
Curifactory Martindale et al., 2022 is a Python API and CLI tool for organizing, tracking, reproducing, and exporting computational research experiments and data analysis workflows. It is intended primarily for smaller teams conducting research, rather than production-level or large-scale ML projects. Curifactory is available on GitHub[1] with an open-source BSD-3-Clause license. Below, we describe the mechanisms within Curifactory to support each of the six capabilities, and compare it with the tools discussed above.
Orchestration¶
Curifactory provides several abstractions, the lowest level of which is a
stage. A stage is a function that takes a defined set of input variable names,
a defined set of output variable names, and an optional set of caching
strategies for the outputs. Stages are similar to Kedro’s nodes but implemented
with @stage() decorators on the target function rather than passing the
target function to a node() call. One level up from a stage is an
experiment: an experiment describes the orchestration of these stages as shown
in Figure 1, functionally chaining them together without
needing to explicitly manage what variables are passed between the stages.
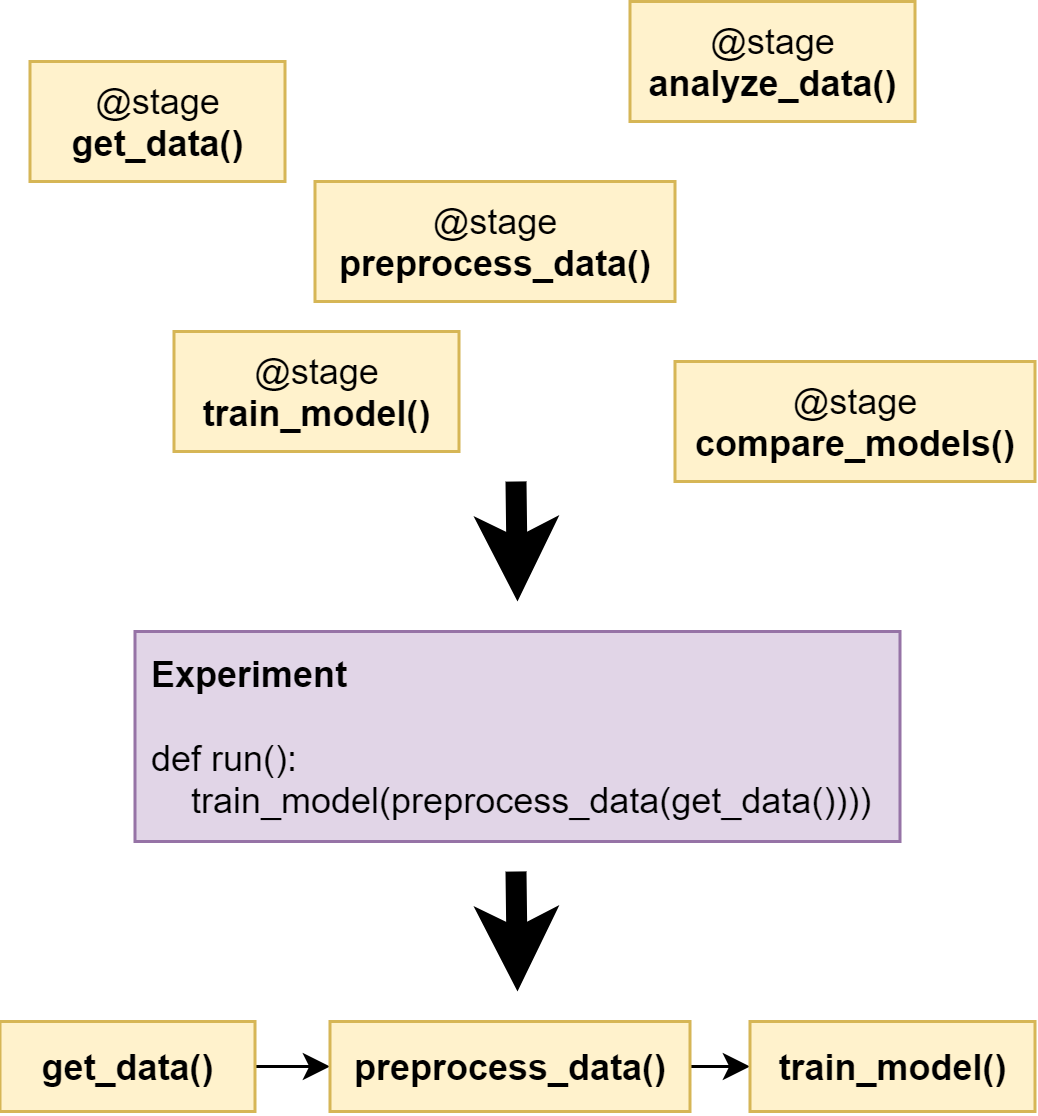
Figure 1:Stages are composed into an experiment.
@stage(inputs=None, outputs=["data"])
def load_data(record):
# every stage has the currently active record
# passed to it, which contains the "state", or
# all previous output values associated with
# the current argset, as defined in the
# Parameterization section
# ...
@stage(inputs=["data"], outputs=["model", "stats"])
def train_model(record, data):
# ...
@stage(inputs=["model"], outputs=["results"])
def test_model(record, model):
# ...
def run(argsets, manager):
"""An example experiment definition.
The primary intent of an experiment is to run
each set of arguments through the desired
stages, in order to compare results at the end.
"""
for argset in argsets:
# A record is the "pipeline state"
# associated with each set of arguments.
# Stages take and return a record,
# automatically handling pushing and
# pulling inputs and outputs from the
# record state.
record = Record(manager, argsets)
test_model(train_model(load_data(record)))Parameterization¶
Parameterization in Curifactory is done directly in Python scripts. The user defines a dataclass with the parameters they need throughout their various stages in order to customize the experiment, and they can then define parameter files that each return one or more instances of this arguments class. All stages in an experiment are automatically given access to the current argument set in use while an experiment is running.
While configuration can also be done directly in Python in Sacred, Curifactory
makes a different trade-off: A parameter file or get_params() function in
Curifactory returns an array of one or more argument sets, and arguments can directly
include complex Python objects. Unlike Sacred, this means Curifactory cannot
directly translate back and forth from static configuration files, but in
exchange allows for grid searches to be defined directly and easily in a single
parameter file, as well as allowing argument sets to be composed or even inherit
from other argument set instances. Importantly, Curifactory can still encode
representations of arguments into JSON for provenance, but this is a one
directional transformation.
This approach allows a great deal of flexibility, and is valuable in experiments where a large range of parameters need to be tested or there is significant repetition among parameter sets. For example, in an experiment testing different effects of model training hyperparameters, there may be several parameter files meant to vary only the arguments needed for model training while using the same base set of data cleaning arguments. Composing these parameter sets from a common imported set means that any subsequent changes to the data cleaning arguments only need to be modified in one place, rather than each individual parameter file.
@dataclass
class MyArgs(curifactory.ExperimentArgs):
"""Define the possible arguments needed in the
stages."""
random_seed: int = 42
train_test_ratio: float = 0.8
layers: tuple = (100,)
activation: str = "relu"
def get_params():
"""Define a simple grid search: return
many arguments instances for testing."""
args = []
layer_sizes = [10, 20, 50, 100]
for size in layer_sizes:
args.append(MyArgs(name=f"network_{size}",
layers=(size,)))
return argsCaching¶
Curifactory supports per-stage caching, similar to memoization, through a set of easy-to-use caching strategies. When a stage executes, it uses the specified cache mechanism to store the stage outputs to disk, with a filename based on the experiment, stage, and a hash of the arguments. When the experiment is re-executed, if it finds an existing output on disk based on this name, it short-circuits the stage computation and simply reloads the previously cached files, allowing a form of re-entrancy. Adding this caching ability to a stage is done through simply providing the list of caching strategies to the stage decorator, one for each output:
@stage(
inputs=["data"],
outputs=["training_set", "testing_set"],
cachers=[PandasCSVCacher]*2
):
def split_data(record, data):
# stage definitionReproducibility¶
As mentioned before, reproducibility consists of tracking provenance and metadata of artifacts as well as providing a means to set up and repeat an experiment in a different compute environment. To handle provenance, Curifactory automatically records metadata for every experiment run executed, including a logfile of the console output, current Git commit hash, argument sets used and the rendered versions of those arguments, and the CLI command used to start the run. The final reports from each run also include a graphical representation of the stage DAG, and shows each output artifact and what its cache file location is.
Curifactory has two mechanisms to fully track and export an experiment run. The first is to execute a “full store” run, which creates a single exported folder containing all metadata mentioned above, along with a copy of every cache file created, the output run report (mentioned below), as well as a Python requirements.txt and Conda environment dump, containing a list of all packages in the environment and their respective versions. This run folder can then be distributed. Reproducing from the folder consists of setting up an environment based on the Conda/Python dependencies as needed, and running the experiment command using the exported folder as the cache directory.
The second mechanism is a command to create a Docker container that includes the environment, entire codebase, and artifact cache for a specific experiment run. Curifactory comes with a default Dockerfile for this purpose, and running the experiment with the Docker flag creates an image that exposes a Jupyter notebook to repeat the run and keep the artifacts in memory, as well as a file server pointing to the appropriate cache for manual exploration and inspection. Directly reproducing the experiment can be done either through the exposed notebook or by running the Curifactory experiment command inside of the image.
Reporting¶
While Curifactory does not run a live web dashboard like MLFlow, DVC’s Iterative Studio, and Kedro-viz, every experiment run outputs an HTML experiment report and updates a top-level index HTML page linking to the new report, which can be browsed from a file manager or statically served if running from an external compute resource. Although simplistic, this reduces the dependencies and infrastructure needed to achieve a basic level of reporting, and produces stand-alone folders for consumption outside of the original environment if needed.
Every report from Curifactory includes all relevant metadata mentioned above, including the machine host name, experiment sequential run number, Git commit hash, parameters, and command line string. Stage code can add user-defined objects to output in each report, such as tables, figures, and so on. Curifactory comes with a default set of helpers for several basic types of output visualizations, including basic line plots, entire Matplotlib figures, and dataframes.
The output report also contains a graphical representation of the DAG for the experiment, rendered using Graphviz, and shows the artifacts produced by each stage and the file path where they are cached. An example of some of the components of this report are rendered in Figure 2, Figure 3, Figure 4, and Figure 5.
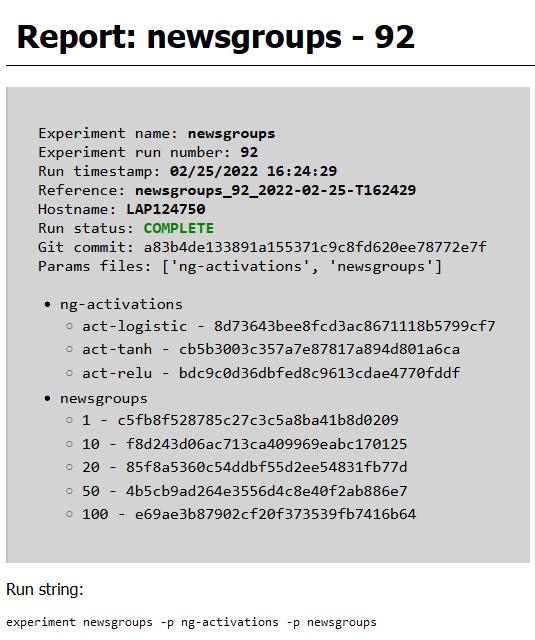
Figure 2:Metadata block at the top of a report.
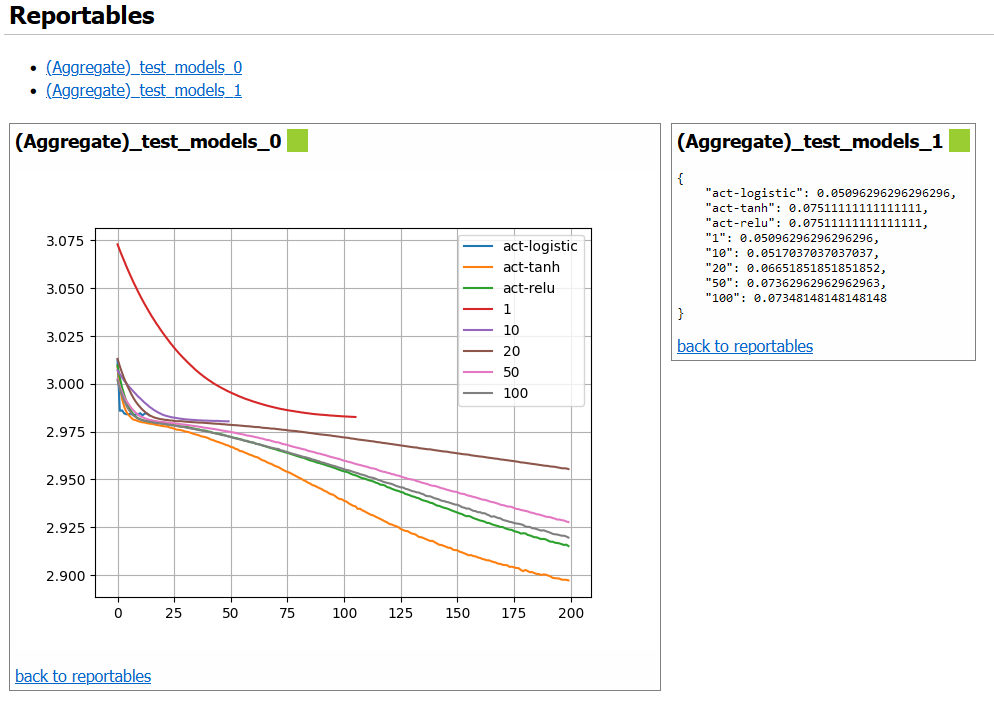
Figure 3:User-defined objects to report (“reportables”).
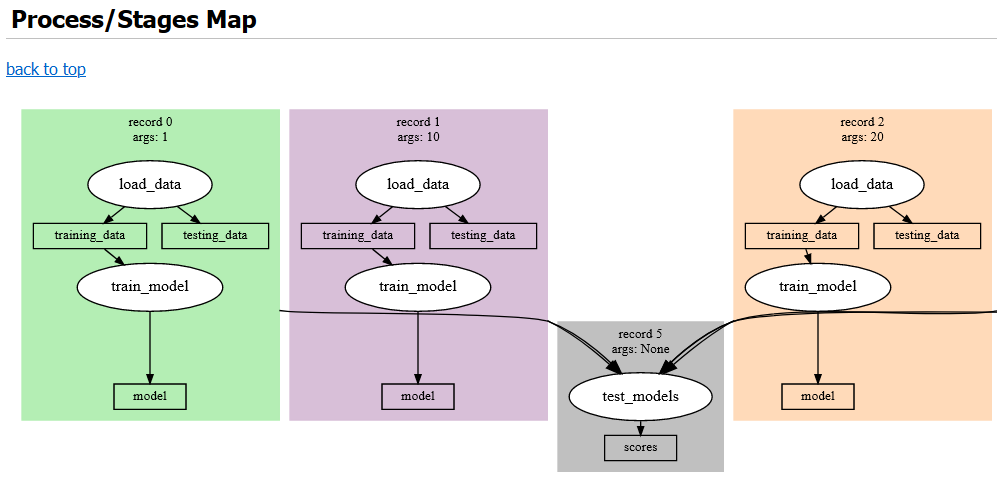
Figure 4:Graphviz rendering of experiment DAG. Each large colored area represents a single record associated with a specific argset. White ellipses are stages, and the blocks in between them are the input and output artifacts.
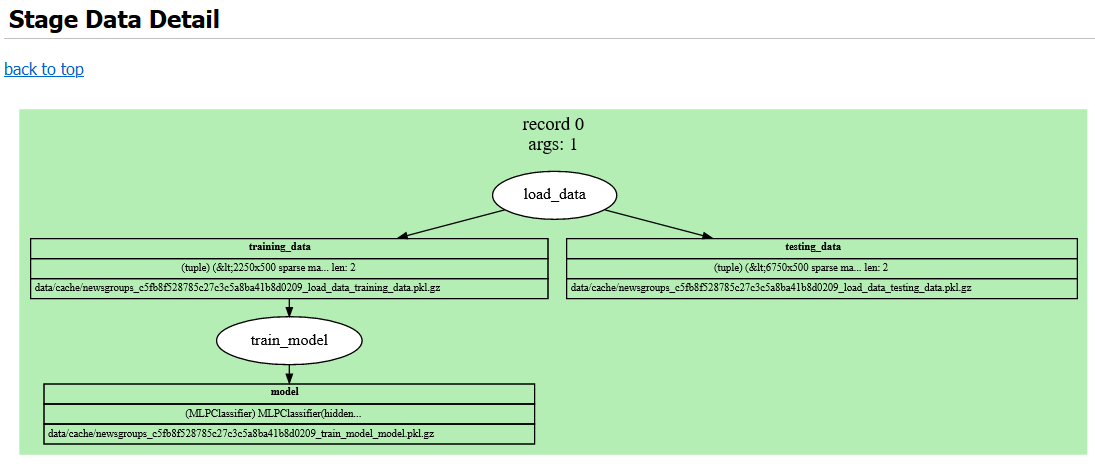
Figure 5:Graphviz rendering of each record in more depth, showing cache file paths and artifact data types.
Scalability¶
Curifactory has no integrated method of executing portions of jobs on external compute resources like Kedro and MLFlow, but it does allow local multi-process parallelization of parameter sets. When an experiment run would entail executing a series of stages for each argument set in series, Curifactory can divide the collection of argument sets into one subcollection per process, and runs the experiment in parallel on each subcollection. By taking advantage of the caching mechanism, when all parallel runs complete, the experiment reruns in a single process to aggregate all of the precached values into a single report.
Conclusion¶
The complexity in modern software, environments, and data analytic approaches threaten the reproducibility and effectiveness of computation-based studies. This has been compounded by the lack of standardization in infrastructure tools and software engineering principles applied within scientific research domains. While many novel tools and systems are in development to address these shortcomings, several design critieria must be met, including the ability to easily compose and orchestrate experiments, parameterize them to manipulate variables under test, cache intermediate artifacts, record provenance of all artifacts and allow the software to port to other systems, produce output visualizations and reports for analysis, and scale execution to the resource requirements of the experiment. We developed Curifactory to address these criteria specifically for small research teams running Python based experiments.
The authors would like to acknowledge the US Department of Energy, National Nuclear Security Administration’s Office of Defense Nuclear Nonproliferation Research and Development (NA-22) for supporting this work.
Oak Ridge National Laboratory This is an open-access article distributed under the terms of the Creative Commons Attribution 3.0 Unported license.
- AI
- artificial intelligence
- API
- application programming interface
- CLI
- command line interface
- DAG
- directed acyclic graph
- DOE
- US Department of Energy
- FAIR
- Findable, Accessible, Interoperable, and Reusable
- HPC
- high-performance computing
- ML
- machine learning
- MLOps
- machine learning operations
- REST
- representational state transfer
- Stodden, V., Borwein, J., & Bailey, D. H. (2013). Publishing Standards for Computational Science: “Setting the Default to Reproducible.” Pennsylvania State University.
- Donoho, D. L., Maleki, A., Rahman, I. U., Shahram, M., & Stodden, V. (2009). Reproducible Research in Computational Harmonic Analysis. Computing in Science Engineering, 11(1), 8–18. 10.1109/MCSE.2009.15
- Hutson, M. (2018). Artificial Intelligence Faces Reproducibility Crisis. Science, 359(6377), 725–726. 10.1126/science.359.6377.725
- Gundersen, O. E., Gil, Y., & Aha, D. W. (2018). On Reproducible AI: Towards Reproducible Research, Open Science, and Digital Scholarship in AI Publications. AI Magazine, 39(3), 56–68. 10.1609/aimag.v39i3.2816
- Storer, T. (2018). Bridging the Chasm: A Survey of Software Engineering Practice in Scientific Programming. ACM Computing Surveys, 50(4), 1–32. 10.1145/3084225
- Peng, R. D. (2011). Reproducible Research in Computational Science. Science, 334(6060), 1226–1227. 10.1126/science.1213847
- Dubois, P. F. (2005). Maintaining Correctness in Scientific Programs. Computing in Science Engineering, 7(3), 80–85. 10.1109/MCSE.2005.54
- Gundersen, O. E., & Kjensmo, S. (2018). State of the Art: Reproducibility in Artificial Intelligence. Proceedings of the AAAI Conference on Artificial Intelligence, 32(1). 10.1609/aaai.v32i1.11503
- Wilkinson, M. D., Dumontier, M., Aalbersberg, Ij. J., Appleton, G., Axton, M., Baak, A., Blomberg, N., Boiten, J.-W., da Silva Santos, L. B., Bourne, P. E., Bouwman, J., Brookes, A. J., Clark, T., Crosas, M., Dillo, I., Dumon, O., Edmunds, S., Evelo, C. T., Finkers, R., … Mons, B. (2016). The FAIR Guiding Principles for Scientific Data Management and Stewardship. Scientific Data, 3(1), 160018. 10.1038/sdata.2016.18
- Lamprecht, A.-L., Garcia, L., Kuzak, M., Martinez, C., Arcila, R., Martin Del Pico, E., Dominguez Del Angel, V., van de Sandt, S., Ison, J., Martinez, P. A., McQuilton, P., Valencia, A., Harrow, J., Psomopoulos, F., Gelpi, J. L., Chue Hong, N., Goble, C., & Capella-Gutierrez, S. (2020). Towards FAIR Principles For Research Software. Data Science, 3(1), 37–59. 10.3233/DS-190026
- Goble, C., Cohen-Boulakia, S., Soiland-Reyes, S., Garijo, D., Gil, Y., Crusoe, M. R., Peters, K., & Schober, D. (2020). FAIR Computational Workflows. Data Intelligence, 2(1–2), 108–121. 10.1162/dint_a_00033
- Mitchell, S. N., Lahiff, A., Cummings, N., Hollocombe, J., Boskamp, B., Reddyhoff, D., Field, R., Zarebski, K., Wilson, A., Burke, M., Archibald, B., Bessell, P., Blackwell, R., Boden, L. A., Brett, A., Brett, S., Dundas, R., Enright, J., Gonzalez-Beltran, A. N., … Reeve, R. (2021). FAIR Data Pipeline: Provenance-Driven Data Management for Traceable Scientific Workflows. arXiv:2110.07117 [Cs, q-Bio].
- Deelman, E., Gannon, D., Shields, M., & Taylor, I. (2009). Workflows and E-Science: An Overview of Workflow System Features and Capabilities. Future Generation Computer Systems, 25, 524–540. 10.1016/j.future.2008.06.012
- Namaki, M. H., Floratou, A., Psallidas, F., Krishnan, S., Agrawal, A., Wu, Y., Zhu, Y., & Weimer, M. (2020). Vamsa: Automated Provenance Tracking in Data Science Scripts. Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, 1542–1551. 10.1145/3394486.3403205
