
Wailord: Parsers and Reproducibility for Quantum Chemistry
Abstract¶
Data driven advances dominate the applied sciences landscape, with quantum chemistry being no exception to the rule. Dataset biases and human error are key bottlenecks in the development of reproducible and generalized insights. At a computational level, we demonstrate how changing the granularity of the abstractions employed in data generation from simulations can aid in reproducible work. In particular, we introduce wailord (https://wailord.xyz), a free-and-open-source python library to shorten the gap between data-analysis and computational chemistry, with a focus on the ORCA suite binaries. A two level hierarchy and exhaustive unit-testing ensure the ability to reproducibly describe and analyze “computational experiments.” wailord offers both input generation, with enhanced analysis, and raw output analysis, for traditionally executed ORCA runs. The design focuses on treating output and input generation in terms of a mini domain specific language instead of more imperative approaches, and we demonstrate how this abstraction facilitates chemical insights.
Introduction¶
The use of computational methods for chemistry is ubiquitous and few modern chemists retain the initial skepticism of the field Kohn, 1999Schaefer, 1986. Machine learning has been further earmarked Meyer et al., 2019Dral, 2020Schütt et al., 2019 as an effective accelerator for computational chemistry at every level, from DFT Gao et al., 2016 to alchemical searches De et al., 2016 and saddle point searches Ásgeirsson & Jónsson, 2018. However, these methods trade technical rigor for vast amounts of data, and so the ability to reproduce results becomes increasingly more important. Independently, the ability to reproduce results Peng, 2011Sandve et al., 2013 in all fields of computational research, and has spawned a veritable flock of methodological and programmatic advances Community et al., 2019, including the sophisticated provenance tracking of AiiDA Pizzi et al., 2016Huber et al., 2020.
Dataset bias¶
Dataset bias Engstrom et al., 2020Blum & Stangl, 2019Rahaman et al., 2019 has gained prominence in the machine learning literature, but has not yet percolated through to the chemical sciences community. At its core, the argument for dataset biases in generic machine learning problems of image and text classification, can be linked to the difficulty in obtaining labeled results for training purposes. This is not an issue in the computational physical sciences at all, as the training data can often be labeled without human intervention. This is especially true when simulations are carried out at varying levels of accuracy. However, this also leads to a heavy reliance on high accuracy calculations on “benchmark” datasets and results Hoja et al., 2021Senior et al., 2019.
Compute is expensive, and the reproduction of data which is openly available is
often hard to justify as a valid scientific endeavor. Rather than focus on the
observable outputs of calculations, instead we assert that it is best to be able
to have reproducible confidence in the elements of the workflow. In the
following sections, we will outline wailord, a library which implements a
two level structure for interacting with ORCA Neese, 2012
to implement an end-to-end workflow to analyze and prepare datasets. Our focus
on ORCA is due to its rapid and responsive development cycles, that it is free
to use (but not open source) and also because of its large repertoire of
computational chemistry calculations. Notably, the black-box nature of ORCA (in
that the source is not available) mirrors that of many other packages (which are
not free) like VASP Hafner, 2008. Using ORCA
then, allows us to design a workflow which is best suited for working with many
software suites in the community.
We shall understand this wailord from the lens of what is often
known as a design pattern in the practice of computational science and
engineering. That is, a template or description to solve commonly occurring
problems in the design of programs.
Structure and Implementation¶
Python has grown to become the lingua-franca for much of the scientific
community
Oliphant, 2007Millman & Aivazis, 2011,
in no small part because of its interactive nature. In particular, the REPL
(read-evaluate-print-loop) structure which has been prioritized (from IPython to
Jupyter) is one of the prime motivations for the use of Python as an exploratory
tool. Additionally, PyPI, the python package index, accelerates the widespread
disambiguation of software packages. Thus wailord is implemented as a free
and open source python library.
Structure¶
Data generation involves set of known configurations (say, xyz inputs) and a
series of common calculations whose outputs are required. Computational
chemistry packages tend to be focused on acceleration and setup details on a
per-job scale. wailord, in contrast, considers the outputs of simulations
to form a tree, where the actual run and its inputs are the leaves, and each
layer of the tree structure holds information which is collated into a single
dataframe which is presented to the user.
Downstream tasks for simulations of chemical systems involve questions phrased
as queries or comparative measures. With that in mind, wailord generates
pandas dataframes which are indistinguishable from standard machine learning
information sources, to trivialize the data-munging and preparation process. The
outputs of wailord represent concrete information and it is not meant to
store runs like the ASE database Larsen et al., 2017 ,
nor run a process to manage discrete workflows like AiiDA
Huber et al., 2020.
By construction, it differs also from existing “interchange” formats as those
favored by the materials data repositories like the QCArchive project
Smith et al., 2021 and is partially close in spirit to the
cclib endeavor O'boyle et al., 2008.
Implementation¶
Two classes form the backbone of the data-harvesting process. The intended point
of interface with a user is the orcaExp class which collects information
from multiple ORCA outputs and produces dataframes which include relevant
metadata (theory, basis, system, etc.) along with the requested results (energy
surfaces, energies, angles, geometries, frequencies, etc.). A lower level “orca
visitor” class is meant to parse each individual ORCA output. Until the release
of ORCA 5 which promises structured property files, the outputs are necessarily
parsed with regular expressions, but validated extensively. The focus on ORCA
has allowed for more exotic helper functions, like the calculation of rate
constants from orcaVis files. However, beyond this functionality offered by
the quantum chemistry software (ORCA), a computational chemistry workflow
requires data to be more malleable. To this end, the plain-text or binary
outputs of quantum chemistry software must be further worked on (post-processed)
to gain insights. This means for example, that the outputs may be entered into a
spreadsheet, or into a plain text note, or a lab notebook, but in practice,
programming languages are a good level of abstraction. Of the programming
languages, Python as a general purpose programming language with a high rate of
community adoption is a good starting place.
Python has a rich set of structures implemented in the standard library, which
have been liberally used for structuring outputs. Furthermore, there have been
efforts to convert the grammar of graphics Wilkinson & Wills, 2005
and tidy-data Wickham et al., 2019 approaches to the pandas
package which have also been adapted internally, including strict unit adherence
using the pint library. The user is not burdened by these implementation
details and is instead ensured a pandas data-frame for all operations, both
at the orcaVis level, and the orcaExp level.
Software industry practices have been followed throughout the development process. In particular, the entire package is written in a test-driven-development (TDD) fashion which has been proven many times over for academia Desai et al., 2008 and industry Bhat & Nagappan, 2006. In essence, each feature is accompanied by a test-case. This is meant to ensure that once the end-user is able to run the test-suite, they are guaranteed the features promised by the software. Additionally, this means that potential bugs can be submitted as a test case which helps isolate errors for fixes. Furthermore, software testing allows for coverage metrics, thereby enhancing user and development confidence in different components of any large code-base.
User Interface¶
The core user interface is depicted in Figure 1. The test suites
cover standard usage and serve as ad-hoc tutorials. Additionally, jupyter
notebooks are also able to effectively run wailord which facilitates its use
over SSH connections to high-performance-computing (HPC) clusters. The user is
able to describe the nature of calculations required in a simple YAML file
format. A command line interface can then be used to generate inputs, or another
YAML file may be passed to describe the paths needed. A very basic harness
script for submissions is also generated which can be rate limited to ensure
optimal runs on an HPC cluster.
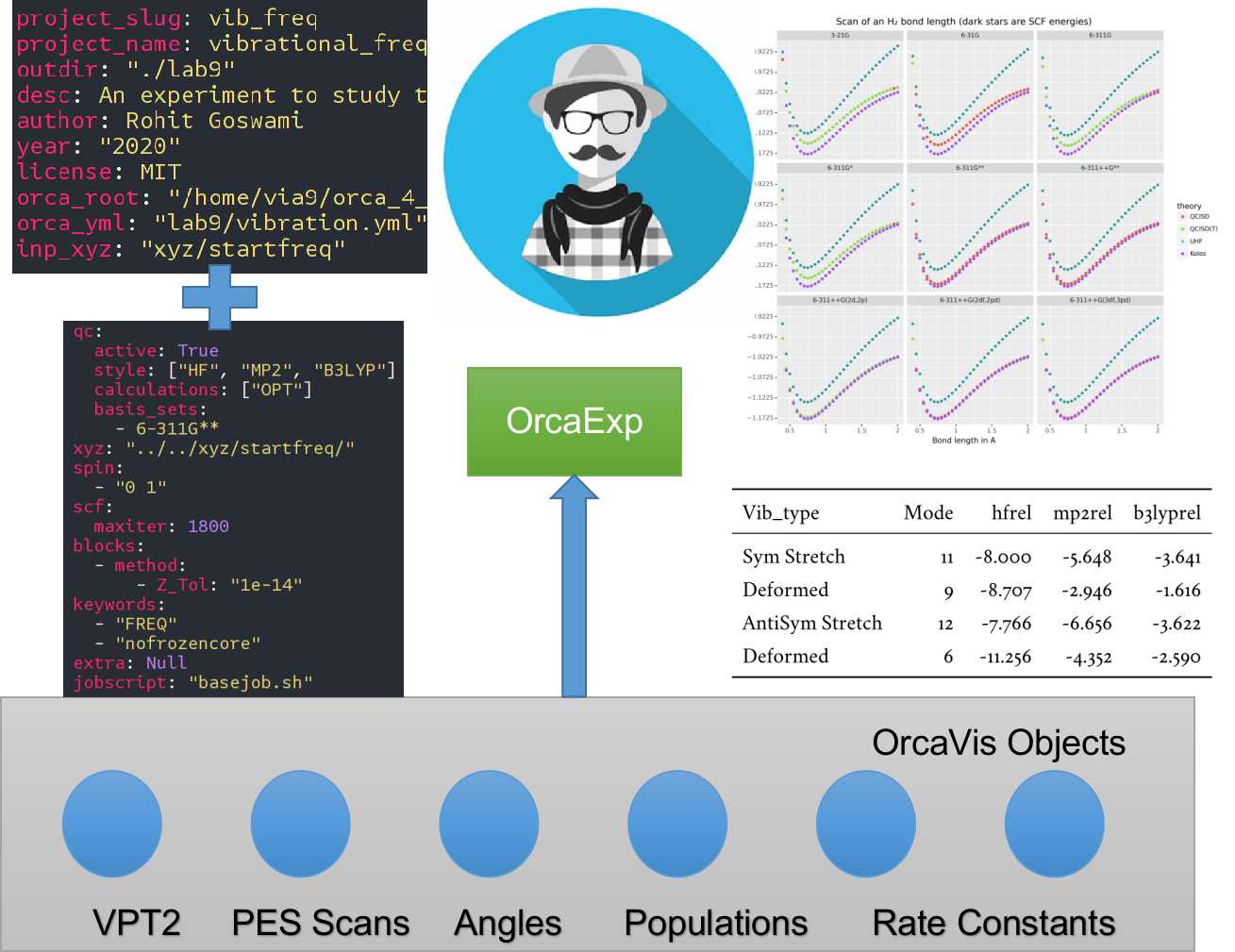
Figure 1:Some implemented workflows including the two input YML files. VPT2 stands for second-order vibrational perturbation theory and Orca_vis objects are part of wailord’s class structure. PES stands for potential energy surface.
Design and Usage¶
A simulation study can be broken into:
- Inputs + Configuration for runs + Data for structures
- Outputs per run
- Post-processing and aggregation
From a software design perspective, it is important to recognize the right level
of abstraction for the given problem. An object-oriented pattern is seen to be
the correct design paradigm. However, though combining test driven development
and object oriented design is robust and extensible, the design of wailord
is meant to tackle the problem at the level of a domain specific language.
Recall from formal language theory Aho & Aho, 2007
the fact that a grammar is essentially meant to specify the entire possible set
of inputs and outputs for a given language. A grammar can be expressed as a
series of tokens (terminal symbols) and non-terminal (syntactic variables)
symbols along with rules defining valid combinations of these.
It may appear that there is little but splitting hairs between parsing data line by line as is traditionally done in libraries, compared to defining the exact structural relations between allowed symbols. However, this design, apart from disallowing invalid inputs, also makes sense from a pedagogical perspective.
For example, of the inputs, structured data like configurations (XYZ formats) are best handled by concrete grammars, where each rule is followed in order:
grammar_xyz = Grammar(
r"""
meta = natoms ws coord_block ws?
natoms = number
coord_block = (aline ws)+
aline = (atype ws cline)
atype = ~"[a-zA-Z]" / ~"[0-9]"
cline = (float ws float ws float)
float = pm number "." number
pm = ~"[+-]?"
number = ~"\\d+"
ws = ~"\\s*"
"""
)This definition maps neatly into the exact specification of an xyz file:
2
H -2.8 2.8 0.1
H -3.2 3.4 0.2Where we recognize that the overarching structure is of the number of atoms,
followed by multiple coordinate blocks followed by optional whitespace. We move
on to define each coordinate block as a line of one or many aline
constructs, each of which is an atype with whitespace and three float values
representing coordinates. Finally we define the positive, negative, numeric and
whitespace symbols to round out the grammar. This is the exact form of every
valid xyz file. The parsimonious library allows handling grammatical
constructs in a Pythonic manner.
However, the generation of inputs is facilitated through the use of generalized
templates for “experiments” controlled by cookiecutter. This allows for
validations on the workflow during setup itself.
For the purposes of the simulation study, one “experiment” consists of multiple single-shot runs; each of which can take a long time.
Concretely, the top-level “experiment” is controlled by a YAML file:
project_slug: methylene
project_name: singlet_triplet_methylene
outdir: './lab6'
desc: An experiment to calculate singlet and triplet states differences at a QCISD(T) level
author: Rohit
year: '2020'
license: MIT
orca_root: '/home/orca/'
orca_yml: 'orcaST_meth.yml'
inp_xyz: 'ch2_631ppg88_trip.xyz'Where each run is then controlled individually.
qc:
active: True
style: ['UHF', 'QCISD', 'QCISD(T)']
calculations: ['OPT']
basis_sets:
- 6-311++G**
xyz: 'inp.xyz'
spin:
- '0 1' # Singlet
- '0 3' # Triplet
extra: '!NUMGRAD'
viz:
molden: True
chemcraft: True
jobscript: 'basejob.sh'Usage is then facilitated by a high-level call.
waex.cookies.gen_base(
template="basicExperiment",
absolute=False,
filen="./lab6/expCookieST_meth.yml",
)The resulting directory tree can be sent to a High Performance Computing Cluster (HPC), and once executed via the generated run-script helper; locally analysis can proceed.
mdat = waio.orca.genEBASet(Path("buildOuts") / \
"methylene",
deci=4)
print(mdat.to_latex(index=False,
caption="CH2 energies and angles \
at various levels of theory, with NUMGRAD"))In certain situations, ordering may be relevant as well (e.g. for generating curves of varying density functional theoretic complexity). This can be handled as well.
For the outputs, similar to the key ideas across signac, nix, spack
and other tools, control is largely taken away from the user in terms of the
auto-generated directory structure. The outputs of each run is largely collected
through regular expressions, due to the ever changing nature of the outputs of
closed source software.
Importantly, for a code which is meant to confer insights, the concept of units
is key. wailord with ORCA has first class support for units using
pint.
Dissociation of H2¶
As a concrete example, we demonstrate a popular pedagogical exercise, namely to obtain the binding energy curves of the H2 molecule at varying basis sets and for the Hartree Fock, along with the results of Kolos & Wolniewicz (1968). We first recognize, that even for a moderate 9 basis sets with 33 points, we expect around 1814 data points. Where each basis set requires a separate run, this is easily expected to be tedious.
Naively, this would require modifying and generating ORCA input files.
!UHF 3-21G ENERGY
%paras
R = 0.4, 2.0, 33 # x-axis of H1
end
*xyz 0 1
H 0.00 0.0000000 0.0000000
H {R} 0.0000000 0.0000000
*We can formulate the requirement imperatively as:
qc:
active: True
style: ['UHF', 'QCISD', 'QCISD(T)']
calculations: ['ENERGY'] # Same as single point or SP
basis_sets:
- 3-21G
- 6-31G
- 6-311G
- 6-311G*
- 6-311G**
- 6-311++G**
- 6-311++G(2d,2p)
- 6-311++G(2df,2pd)
- 6-311++G(3df,3pd)
xyz: 'inp.xyz'
spin:
- '0 1'
params:
- name: R
range: [0.4, 2.00]
points: 33
slot:
xyz: True
atype: 'H'
anum: 1 # Start from 0
axis: 'x'
extra: Null
jobscript: 'basejob.sh'This run configuration is coupled with an experiment setup file, similar to the one in the previous section. With this in place, generating a data-set of all the required data is fairly trivial.
kolos = pd.read_csv(
"../kolos_H2.ene",
skiprows=4,
header=None,
names=["bond_length", "Actual Energy"],
sep=" ",
)
kolos['theory']="Kolos"
expt = waio.orca.orcaExp(expfolder=Path("buildOuts") / "h2")
h2dat = expt.get_energy_surface()Finally, the resulting data can be plotted using tidy principles.
imgname = "images/plotH2A.png"
p1a = (
p9.ggplot(
data=h2dat, mapping=p9.aes(x="bond_length",
y="Actual Energy",
color="theory")
)
+ p9.geom_point()
+ p9.geom_point(mapping=p9.aes(x="bond_length",
y="SCF Energy"),
color="black", alpha=0.1,
shape='*', show_legend=True)
+ p9.geom_point(mapping=p9.aes(x="bond_length",
y="Actual Energy",
color="theory"),
data=kolos,
show_legend=True)
+ p9.scales.scale_y_continuous(breaks
= np.arange( h2dat["Actual Energy"].min(),
h2dat["Actual Energy"].max(), 0.05) )
+ p9.ggtitle("Scan of an H2 \
bond length (dark stars are SCF energies)")
+ p9.labels.xlab("Bond length in Angstrom")
+ p9.labels.ylab("Actual Energy (Hatree)")
+ p9.facet_wrap("basis")
)
p1a.save(imgname, width=10, height=10, dpi=300)Which gives rise to the concise representation Figure 2 from which all required inference can be drawn.
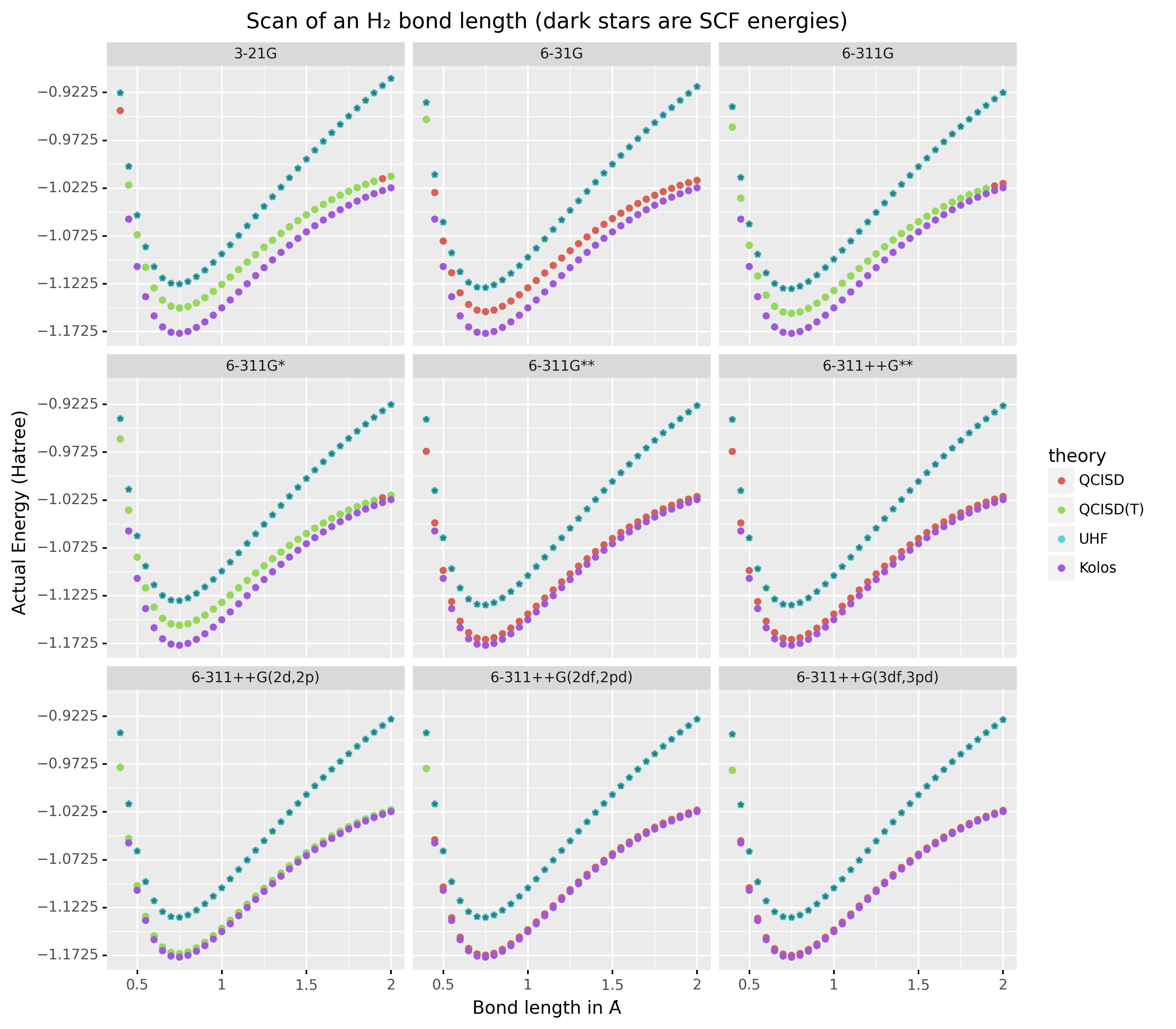
Figure 2:Plots generated from tidy principles for post-processing wailord parsed outputs.
In this particular case, it is possible to see the deviations from the experimental results at varying levels of theory for different basis sets.
Conclusions¶
We have discussed wailord in the context of generating, in a reproducible
manner the structured inputs and output datasets which facilitate chemical
insight. The formulation of bespoke datasets tailored to the study of specific
properties across a wide range of materials at varying levels of theory has been
shown. The test-driven-development approach is a robust methodology for
interacting with closed source software. The design patterns expressed, of
which the wailord library is a concrete implementation, is expected to be
augmented with more workflows, in particular, with a focus on nudged elastic
band. The methodology here has been applied to ORCA, however, the two level
structure has generalizations to most quantum chemistry codes as well.
Importantly, we note that the ideas expressed form a design pattern for interacting with a plethora of computational tools in a reproducible manner. By defining appropriate scopes for our structured parsers, generating deterministic directory trees, along with a judicious use of regular expressions for output data harvesting, we are able to leverage tidy-data principles to analyze the results of a large number of single-shot runs.
Taken together, this tool-set and methodology can be used to generate elegant reports combining code and concepts together in a seamless whole. Beyond this, the interpretation of each computational experiment in terms of a concrete domain specific language is expected to reduce the requirement of having to re-run benchmark calculations.
R Goswami thanks H. Jónsson and V. Ásgeirsson for discussions on the design of computational experiments for inference in computation chemistry. This work was partially supported by the Icelandic Research Fund, grant number 217436052.
Copyright © 2022 Goswami. This is an open-access article distributed under the terms of the Creative Commons Attribution 3.0 Unported license.
- HPC
- high-performance-computing
- PES
- potential energy surface
- TDD
- test-driven-development
- Kohn, W. (1999). Nobel Lecture: Electronic Structure of Matter—Wave Functions and Density Functionals. Reviews of Modern Physics, 71(5), 1253–1266. 10.1103/RevModPhys.71.1253
- Schaefer, H. F. (1986). Methylene: A Paradigm for Computational Quantum Chemistry. Science, 231(4742), 1100–1107. 10.1126/science.231.4742.1100
- Meyer, R., Schmuck, K. S., & Hauser, A. W. (2019). Machine Learning in Computational Chemistry: An Evaluation of Method Performance for Nudged Elastic Band Calculations. Journal of Chemical Theory and Computation, 15(11), 6513–6523. 10.1021/acs.jctc.9b00708
- Dral, P. O. (2020). Quantum Chemistry in the Age of Machine Learning. The Journal of Physical Chemistry Letters, 11(6), 2336–2347. 10.1021/acs.jpclett.9b03664
- Schütt, K. T., Gastegger, M., Tkatchenko, A., Müller, K.-R., & Maurer, R. J. (2019). Unifying Machine Learning and Quantum Chemistry with a Deep Neural Network for Molecular Wavefunctions. Nature Communications, 10(1), 5024. 10.1038/s41467-019-12875-2
- Gao, T., Li, H., Li, W., Li, L., Fang, C., Li, H., Hu, L., Lu, Y., & Su, Z.-M. (2016). A Machine Learning Correction for DFT Non-Covalent Interactions Based on the S22, S66 and X40 Benchmark Databases. Journal of Cheminformatics, 8(1), 24. 10.1186/s13321-016-0133-7
- De, S., Bartók, A. P., Csányi, G., & Ceriotti, M. (2016). Comparing Molecules and Solids across Structural and Alchemical Space. Physical Chemistry Chemical Physics, 18(20), 13754–13769. 10.1039/C6CP00415F
- Ásgeirsson, V., & Jónsson, H. (2018). Exploring Potential Energy Surfaces with Saddle Point Searches. In W. Andreoni & S. Yip (Eds.), Handbook of Materials Modeling (pp. 1–26). Springer International Publishing. 10.1007/978-3-319-42913-7_28-1
- Peng, R. D. (2011). Reproducible Research in Computational Science. Science, 334(6060), 1226–1227.
- Sandve, G. K., Nekrutenko, A., Taylor, J., & Hovig, E. (2013). Ten Simple Rules for Reproducible Computational Research. PLOS Computational Biology, 9(10), e1003285.
- Community, T. T. W., Arnold, B., Bowler, L., Gibson, S., Herterich, P., Higman, R., Krystalli, A., Morley, A., O’Reilly, M., & Whitaker, K. (2019). The Turing Way: A Handbook for Reproducible Data Science. Zenodo.
- Pizzi, G., Cepellotti, A., Sabatini, R., Marzari, N., & Kozinsky, B. (2016). AiiDA: Automated Interactive Infrastructure and Database for Computational Science. Computational Materials Science, 111, 218–230. 10.1016/j.commatsci.2015.09.013
- Huber, S. P., Zoupanos, S., Uhrin, M., Talirz, L., Kahle, L., Häuselmann, R., Gresch, D., Müller, T., Yakutovich, A. V., Andersen, C. W., Ramirez, F. F., Adorf, C. S., Gargiulo, F., Kumbhar, S., Passaro, E., Johnston, C., Merkys, A., Cepellotti, A., Mounet, N., … Pizzi, G. (2020). AiiDA 1.0, a Scalable Computational Infrastructure for Automated Reproducible Workflows and Data Provenance. Scientific Data, 7(1), 300. 10.1038/s41597-020-00638-4
- Engstrom, L., Ilyas, A., Santurkar, S., Tsipras, D., Steinhardt, J., & Madry, A. (2020). Identifying Statistical Bias in Dataset Replication. arXiv:2005.09619 [Cs, Stat].
- Blum, A., & Stangl, K. (2019). Recovering from Biased Data: Can Fairness Constraints Improve Accuracy? arXiv:1912.01094 [Cs, Stat].
